Trying to reduce the memory overhead when using mclapply
I am currently trying to understand how to reduce the memory used by mclapply. This function is rather complicated and others have explained the differences versus parLapply (A_Skelton73, 2013; lockedoff, 2012 ) and also made it clear that in mclapply each job does not know if the others are running out of memory and thus cannot trigger gc (Urbanek, 2012).
While I still struggle to understand all the details of mclapply, I can successfully use it to reduce computation time at the expense of a very high memory load. I am still looking for tips on how to reduce this memory load.
Here is what I have done.
Problem setting
I have a large data set on the form of a data.frame. I want to apply a function that works using subsets of the data.frame without the need for communication between the chunks, and I want to apply the function fast. In other words, I can safely split the matrix and speed the computation process using mclapply.
While this works, I would like to minimize memory consumption.
Toy data
Here is just some toy data for the example.
## The real data set is much larger than this
set.seed(20131113)
data <- data.frame(matrix(rnorm(1e+05), ncol = 10))
dim(data)
## [1] 10000 10
Approach 1
The first approach I have used is to pre-split the data and then use mclapply over the split data. For illustrative purposes, lets say that the function I want to apply is just rowMeans.
## Pre-split the data
dataSplit <- split(data, rep(1:10, each = 1000))
## Approach 1
library("parallel")
res1 <- mclapply(dataSplit, rowMeans, mc.cores = 2)
This gets the job done, but because my real dataSplit is much larger in memory, using say 8-10 cores blows up the memory.
Best way to pre-split?
If I know that if I am using \( n \) number of cores (in this example \( n=2 \) ) and the data set has \( m \) rows, then one option for approach #1 is to split the data into \( n \) chunks each of size \( m / n \) (rounding if needed).
## Pre-split the data into m/n chunks
dataSplit1b <- split(data, rep(1:2, each = 5000))
## Approach 1b
res1b <- mclapply(dataSplit1b, rowMeans, mc.cores = 2)
The memory needed is then in part determined by the chunksize (1000 vs 5000 shown above). One excellent suggestion (via Ben) is to reduce the memory load using this approach is to just smaller chunks. However, the runtime of the function I want to apply (rowMeans in the example) is not very sensible to the chunksize used, thus using very small chunks is not ideal as it increases computation time. Finding the sweet point is tricky, but using chunksizes of \(m / (2n) \) could certainly help memory wise without majorly affecting computation time.
Approach 2
One suggestion (via Roger) is to use an environment in order to minimize copying (and thus memory load) while using mclapply over a set of indexes.
## Save the split data in an environment
my.env <- new.env()
my.env$data1 <- dataSplit1b[[1]]
my.env$data2 <- dataSplit1b[[2]]
## Function that takes indexes, then extracts the data from the environment
applyMyFun <- function(idx, env) {
eval(parse(text = paste0("result <- env$", ls(env)[idx])))
rowMeans(result)
}
## Approach 2
index <- 1:2
names(index) <- 1:2
res2 <- mclapply(index, applyMyFun, env = my.env, mc.cores = 2)
## Same result?
identical(res1b, res2)
## [1] TRUE
Approach 3
Another suggestion (via Roger) is to save the data chunks and load them individually inside the function that I pass to mclapply. This does not seem ideal in terms of having to create the temporary chunk data files. But I would expect this method to have the lowest memory footprint.
## Save the chunks
for (i in names(dataSplit1b)) {
chunk <- dataSplit1b[[i]]
output <- paste0("chunk", i, ".Rdata")
save(chunk, file = output)
}
## Function that loads the chunk
applyMyFun2 <- function(idx) {
load(paste0("chunk", idx, ".Rdata"))
rowMeans(chunk)
}
## Approach 3
res3 <- mclapply(index, applyMyFun2, mc.cores = 2)
## Same result?
identical(res1b, res3)
## [1] TRUE
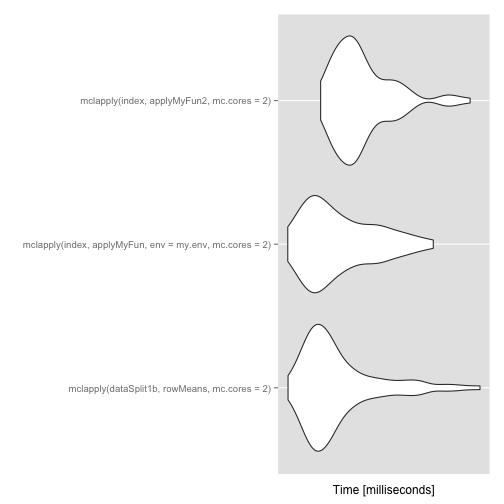
Computation time comparison
Computation time wise, approaches 2 and 3 do not seem very different. Approach 1b seems a tiny bit faster. [Edit: the order of the best approach might change slightly if you re-run this code]
library("microbenchmark")
micro <- microbenchmark(mclapply(dataSplit1b, rowMeans, mc.cores = 2), mclapply(index,
applyMyFun, env = my.env, mc.cores = 2), mclapply(index, applyMyFun2, mc.cores = 2))
micro
## Unit: milliseconds
## expr min lq
## mclapply(dataSplit1b, rowMeans, mc.cores = 2) 17.43 19.97
## mclapply(index, applyMyFun, env = my.env, mc.cores = 2) 17.05 19.20
## mclapply(index, applyMyFun2, mc.cores = 2) 17.19 23.11
## median uq max neval
## 21.41 26.00 65.53 100
## 20.60 23.92 43.67 100
## 24.56 28.39 46.99 100
library("ggplot2")
autoplot(micro)
Memory wise comparison
Relying on the cluster tools for calculating the maximum memory used, I ran each approach (1b, 2, and 3) ten times each using 2 cores using the scripts available in this gist. The maximum memory used showed no variability (within an approach) and the results are that approach 1b used 1.224G RAM, approach 2 used 1.176G RAM, and approach 3 used 1.177G RAM. Not a huge difference. Due to having to write and then load, approach 3 was slower than the other two.
Re-doing the previous test but using 20 cores lead to very similar wall clock computation times between all three approaches and to approaches 1b and 2 for 2 cores. This is due to the nature of the example, aka rowMeans is fast even with the larger chunks. Approach 1b used 7.728G RAM, approach 2 used 7.674G RAM, and approach 3 used 7.690G RAM. Hm…
Using 20 cores with previously created data files (either the split data for approaches 1b and 2, or the chunk files for approach 3) has a very different memory footprint. Approach 1b used in average 6.0744G RAM, approach 2 used 4.2647G RAM , and approach 3 used 2.6545G RAM.
Edit
Ryan from (Ryan 2013) contributed a fourth approach which used 6.794G RAM when starting from scratch with 20 cores. This approach definitely beats the other ones under the condition of starting from scratch. Note that just creating the data object uses 558.938M RAM: multiplied by 20 it would be around 10.92G RAM.
Conclusions
Using 2 or 20 cores, approach 2 beat by a very small margin approaches 3 and 1b in terms of memory usage. However, all approaches failed in terms of not having the memory blow up as you increase the number of cores when starting from scratch.
If a lower memory option is used for splitting the data and creating the chunk files, approach 3 seems like the winner in terms of memory usage. So in pure terms of lowering the memory load on mclapply approach 3 wins, although you still need to create the chunk files and do so without much memory usage.
If you have any ideas or suggestions, please let me know! Thank you!
References
Citations made with knitcitations (Boettiger, 2013).
- A_Skelton73, (2013) understanding the differences between mclapply and parLapply in R. understanding the differences between mclapply and parLapply in R - Stack Overflow http://stackoverflow.com/questions/17196261/understanding-the-differences-between-mclapply-and-parlapply-in-r
- lockedoff, (2012) Using mclapply, foreach, or something else in [r] to operate on an object in parallel?. Using mclapply, foreach, or something else in [r] to operate on an object in parallel? - Stack Overflow http://stackoverflow.com/questions/11036702/using-mclapply-foreach-or-something-else-in-r-to-operate-on-an-object-in-par
- [R-sig-hpc] mclapply: rm intermediate objects and returning memory . https://mailman.stat.ethz.ch/pipermail/r-sig-hpc/2012-October/001534.html
- [Bioc-devel] Trying to reduce the memory overhead when using mclapply . https://stat.ethz.ch/pipermail/bioc-devel/2013-November/004930.html
- Carl Boettiger, (2013) knitcitations: Citations for knitr markdown files. http://CRAN.R-project.org/package=knitcitations
Reproducibility
sessionInfo()
## R version 3.0.2 (2013-09-25)
## Platform: x86_64-apple-darwin10.8.0 (64-bit)
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] parallel stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] microbenchmark_1.3-0 ggplot2_0.9.3.1 knitcitations_0.4-7
## [4] bibtex_0.3-6 knitr_1.5
##
## loaded via a namespace (and not attached):
## [1] codetools_0.2-8 colorspace_1.2-4 dichromat_2.0-0
## [4] digest_0.6.4 evaluate_0.5.1 formatR_0.10
## [7] grid_3.0.2 gtable_0.1.2 httr_0.2
## [10] labeling_0.2 MASS_7.3-29 munsell_0.4.2
## [13] plyr_1.8 proto_0.3-10 RColorBrewer_1.0-5
## [16] RCurl_1.95-4.1 reshape2_1.2.2 scales_0.2.3
## [19] stringr_0.6.2 tools_3.0.2 XML_3.95-0.2
## [22] xtable_1.7-1
Scripts
The scripts are available in this gist. The main one is testApproach.R while the other ones are just job-submitters.
Check other topics on #rstats.