Easily explore a table with shinycsv
Have you ever had to explore a table with data? I believe the answer is yes for most people that work at a computer or even just use it for communicating with their friends and family. Tables of data pop up everywhere, for example in personal finance. Websites like Mint.com allow you to download your transactions in a CSV file called transactions.csv. CSV is one of the many formats for storing tables and most likely when you try to open the transactions.csv file, it will open with Excel. Now, can you make a quick figure of one of your columns in your table?
Some will answer yes, others no. The basic issue is that it’s not super easy to explore your data in Excel or similar programs. Wait, shouldn’t it be easy? 😕
What if you want to subset your data and want to re-make the plot? How about getting some simple statistics like the mean or frequency of some categories for a given variable? 😨 These are some of the immediate tasks that are helpful when exploring data. Visually, making figures with two variables is also very common.
Programmers and experts in Excel, Stata, R among other options can perform these data explorations. It might take them a little bit of time to write the code or remember it or use the user interface menu of their program of choice. But what about everyone else?
At the Lieber Institute for Brain Development where I work, it’s common for us to exchange data in tables, and thus explore data. That’s why we created shinycsv (Collado-Torres, Semick, and Jaffe, 2016). It’s an R package (R Core Team, 2016) that contains a shiny (Chang, Cheng, Allaire, Xie, et al., 2017) application that allows users to interactively explore a table.
Installing R is a pretty high bar, that’s why we are hosting this application at https://jhubiostatistics.shinyapps.io/shinycsv/. Try it out!
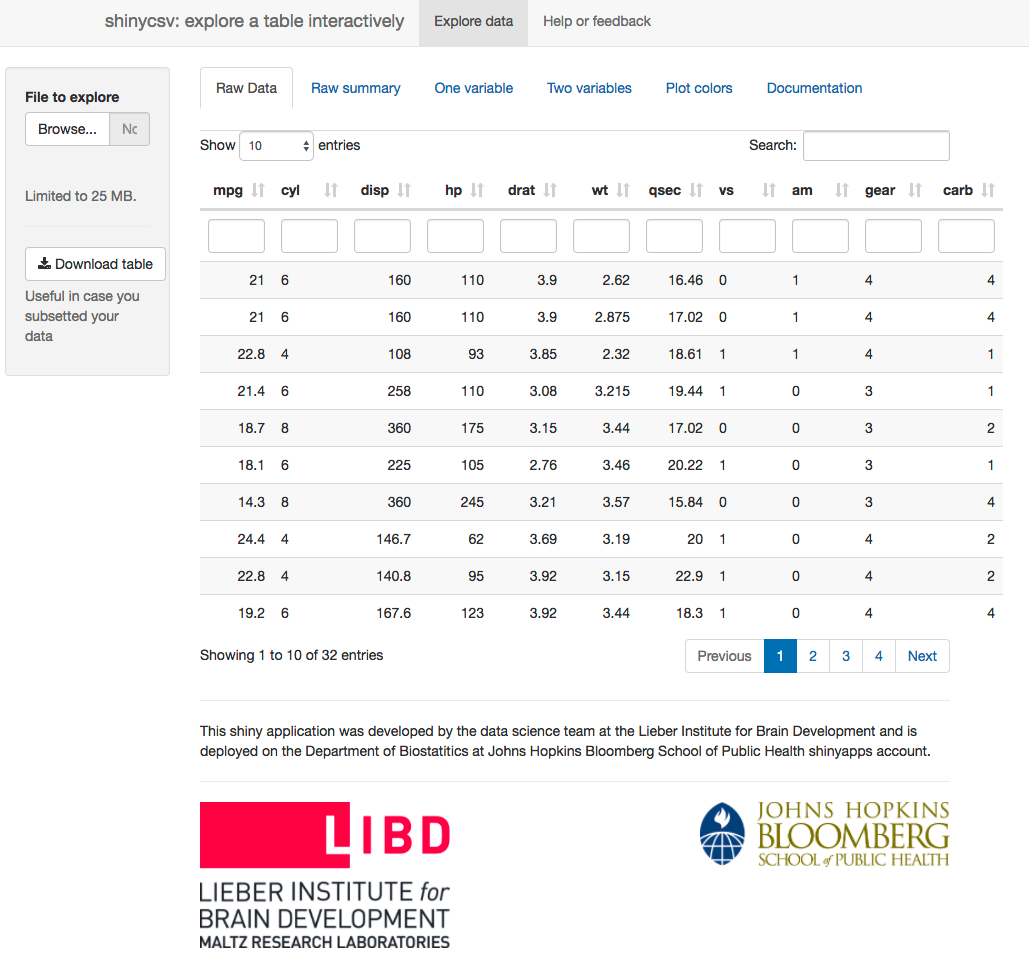
shinycsv application
The application includes data about cars to demonstrate what it can do. It’s a small data set that is commonly used for demonstration purposes. Anyhow, in the application you’ll notice a few tabs.
The application shows the raw data in an interactive table that allows you to subset the observations by some criteria, search in the table, and sort in different ways. The raw summary tab shows quick statistical summaries which depend on the variable type (numerical, categorical, etc). If you interacted with the table in raw data then the summaries at raw summary will be based on the subset you selected.
The one variable and two variables tabs are for making figures based on one or two variables at a time. The code in shinycsv tries to guess what’s the best figure for a given type of variable and in case that you are interested in learning R, it also shows the exact code you can use to reproduce the figure in your computer. We added this feature to excite users about learning R. And it’s useful for advanced users too that might want to customize the resulting figures. Hm…, you don’t like the colors we chose for the figure? Well go to plot colors, choose another color, and come back to see your new figure with the color of your choosing. 😄
Hm… but what if you don’t have a CSV file? Well, shinycsv can handle many different tables thanks to rio (Chan, Chan, Leeper, and Becker, 2016). Even Excel sheets! 😉
So, go ahead and test it out! We’ll be glad to hear your feedback at LieberInstitute/shinycsv.
Notes
- Note that when I referred to tables earlier, I referred to square tables with different variables (age, height, weight, etc) as columns as observations as rows. That is, Excel files with a single sheet with no comments or figures inside the Excel file.
- Are you interested in learning more about
Randshiny? Maybe you’ll want to take a look at the showcase mode version of the application. - If you use
shinycsv::explore()locally, the file size limit is increased to 500 MB. Although at that point you might want to consider usingRor another programming language. - What about casting variables? If you want to have fine control about casting the variables, save your data in a RData file. Sure, this requires an R user.
Reproducibility
## Reproducibility info
library('devtools')
options(width = 120)
session_info()
## Session info -----------------------------------------------------------------------------------------------------------
## setting value
## version R Under development (unstable) (2016-10-26 r71594)
## system x86_64, darwin13.4.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## tz America/New_York
## date 2017-01-20
## Packages ---------------------------------------------------------------------------------------------------------------
## package * version date source
## bibtex 0.4.0 2014-12-31 CRAN (R 3.4.0)
## bitops 1.0-6 2013-08-17 CRAN (R 3.4.0)
## devtools * 1.12.0 2016-12-05 CRAN (R 3.4.0)
## digest 0.6.11 2017-01-03 CRAN (R 3.4.0)
## evaluate 0.10 2016-10-11 CRAN (R 3.4.0)
## httr 1.2.1 2016-07-03 CRAN (R 3.4.0)
## knitcitations * 1.0.7 2015-10-28 CRAN (R 3.4.0)
## knitr * 1.15.1 2016-11-22 CRAN (R 3.4.0)
## lubridate 1.6.0 2016-09-13 CRAN (R 3.4.0)
## magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
## memoise 1.0.0 2016-01-29 CRAN (R 3.4.0)
## plyr 1.8.4 2016-06-08 CRAN (R 3.4.0)
## R6 2.2.0 2016-10-05 CRAN (R 3.4.0)
## Rcpp 0.12.9 2017-01-14 CRAN (R 3.4.0)
## RCurl 1.95-4.8 2016-03-01 CRAN (R 3.4.0)
## RefManageR 0.13.1 2016-11-13 CRAN (R 3.4.0)
## RJSONIO 1.3-0 2014-07-28 CRAN (R 3.4.0)
## stringi 1.1.2 2016-10-01 CRAN (R 3.4.0)
## stringr 1.1.0 2016-08-19 CRAN (R 3.4.0)
## withr 1.0.2 2016-06-20 CRAN (R 3.4.0)
## XML 3.98-1.5 2016-11-10 CRAN (R 3.4.0)
References
Citations made with knitcitations (Boettiger, 2015).
[1] C. Boettiger. knitcitations: Citations for 'Knitr' Markdown Files. R package version 1.0.7. 2015. URL: https://CRAN.R-project.org/package=knitcitations.
[2] C. Chan, G. C. Chan, T. J. Leeper and J. Becker. rio: A Swiss-army knife for data file I/O. R package version 0.4.16. 2016.
[3] W. Chang, J. Cheng, J. Allaire, Y. Xie, et al. shiny: Web Application Framework for R. R package version 1.0.0. 2017. URL: https://CRAN.R-project.org/package=shiny.
[4] L. Collado-Torres, S. Semick and A. E. Jaffe. shinycsv: Explore a table interactively in a shiny application. R package version 0.99.7. 2016. URL: https://github.com/LieberInstitute/shinycsv.
[5] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2016. URL: https://www.R-project.org/.
Want more?
Check other @jhubiostat student blogs at Bmore Biostats as well as topics on #rstats.