Learning from our search history
Origin of the idea
Recently the team I work with has had a few new members and I’ve been thinking lately of ways we could try to help them. The team leader was traveling this week, which gave me the opportunity to come up with a new type of session and test it out. That’s the origin of this learning from our search history idea. We tested it today and I’m quite happy with the results so far, so I thought it would be useful to document what we did and share it with others.
Motivation
The theory
As I show in the slides below, in our group we follow the you must try, and then you must ask framework although with a little different interpretation. The basic goal is to search independently for a period of time (say 15 minutes), but then ask others for help if you are still stuck beyond that point.
In other words, you have to be able to find some answers yourself since that’s part of our job using resources that range from Google Search, to the Bioconductor Support, to the RStudio Community, among other websites. However, we also acknowledge that some questions have difficult answers. Maybe a Stack Overflow thread has multiple answers and not necessarily the top voted question has the answer you are looking for. So instead of spending too much energy, we also tell our members to avoid getting into a rabbit hole for hours. That’s where asking for help comes to play. And you can ask for help from any community you belong to: those involved in the project through Slack, our full team, other scientists in our university, communities we belong to like the rstats community on Twitter, the Bioconductor users community at large, etc.
I did mention that it’s ideal to think about the person who will be helping you answer your question. Small reproducible examples, versions of the software you used, sharing your code on GitHub 1, the commands you used and the order you used them in can all be valuable for resolving different types of problems. Jenny Bryan has talked much more about this subject, for example in this 2018 webinar.
In practice
Trying for a while then asking for help is all good in practice. However, asking for help is very challenging. It’s scary, you open yourself because you show what you don’t know to other people, and sadly historically many questions have been met with negative feedback on the Internet. Thus, they can make you feel dumb, stupid and many other negative emotions.
I do think that asking for help can be worth it and even wrote a previous blog post on this subject. Some reasons why it’s worth it include being able to move on with your work 2, you might learn something new, and if you follow the strategies for helping others help you, you might even figure out the answer yourself.
Now, we all ask for help regardless of how long we have been writing code. Here’s an example tweet that conveys the same message. There are thousands of such tweets online.
I conduct approximately 3948234 programming-related Google searches per day. So does every other developer I know, whether they have 5 weeks of experience or 5 years. Help normalize this practice by tweeting your daily searches with #devgoogle! https://t.co/XfNVUZlhC5
— Lyzi Diamond (@lyzidiamond) January 15, 2019
We are diverse

The team (shown as of May 2018) we work at is very diverse because we all:
- have different backgrounds,
- have acquired different skills,
- are at different career stages (from rotation student up to associate professor),
- have different interests,
- use different operating systems (from Fedora to Ubuntu to macOS to Windows)
- use different tools (mobaxterm vs putty, TextMate vs RStudio, …).
But also because we seek help in different ways 3 and we learn differently.
This means that we have a lot to learn from each other 😊🤓.
Learn from each other exercise
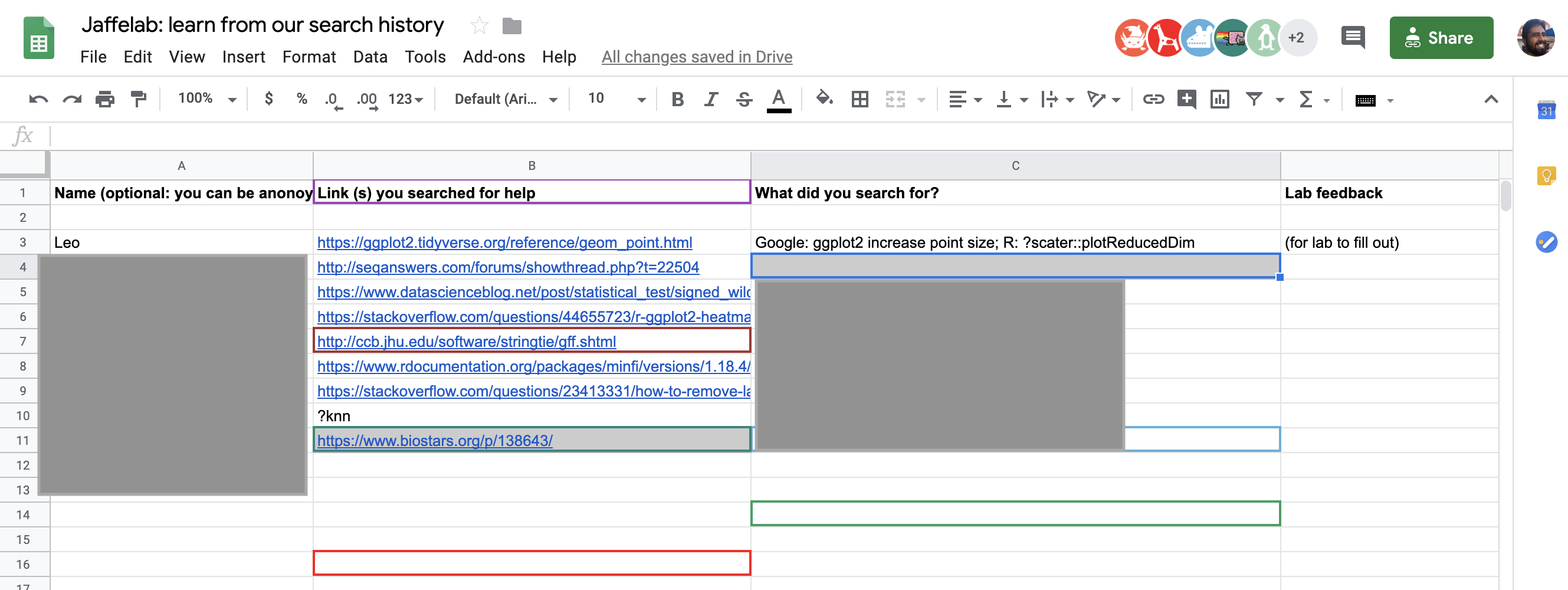
At bit.ly/learnfromsearch you can find a copy of the Google Spreadsheet with the information you need for your team.
Some rules
First, we need to make sure that everyone will feel save to ask questions. That’s why I:
- reminded others about our code of conduct,
- invited everyone to practice their empathy and be mindful that language matters 4,
- to keep everyone’s time in their mind as a question could lead to a longer discussion which is best to take another occasion. 5
Main steps
The idea is that you pick a problem you solved recently and share:
- what you were trying to solve,
- the actual text you searched for in Google or elsewhere 6,
- the link of the website where you find your answer or that guided in this process.
We improved the steps as we were testing this! 🙌🏽🙂
Once everyone has contributed their information to the spreadsheet, we then proceed to go around the table showing and explaining our search use cases.
Goals
Ultimately the goals of this exercise are to
- empower ourselves with the knowledge from our teammates,
- learn from how we all find help,
- build a supportive environment where we feel comfortable asking for help.
Thus in the end, we are enabling our team to fully follow the you must try, and then you must ask framework.
Test session
The first and only session so far went approximately like this:
- Min 0-5: get settled in the room.
- Min 5-22: presentation about the idea to get members to buy into it.
- Min 22-26: demonstration.
- Min 26-33: members prepared their information to share with the team.
- Min 33-58: members presented their problems, the searches the did, and the solution(s) they found.
- Min 58-60: quick wrap up.
My search example
At bit.ly/learnfromsearch I left some examples (anonymized). Mine was about increasing the point size of the output of a plot made with scater::plotReducedDim() which returns a ggplot2 (Wickham, 2016) object. Hence why I searched in Google for ggplot2 increase point size which lead me to the geom_point() reference website. I then tried using + geom_point(size = 20) but that broke the color mapping. I was about to dive into the GitHub code for scater (McCarthy, Campbell, Lun, and Willis, 2017) as this my go-to behavior for many similar quests, but I decided to check the help page using ?scater::plotReducedDim.
… Additional arguments for visualization, see ?“scater-plot-args” for details.
The documentation for ... lead me to ?"scater-plot-args" where I finally found the point_size argument
point_size: Numeric scalar, specifying the size of the points. Defaults to NULL.
and that solved my problem.
So what used to look like this:
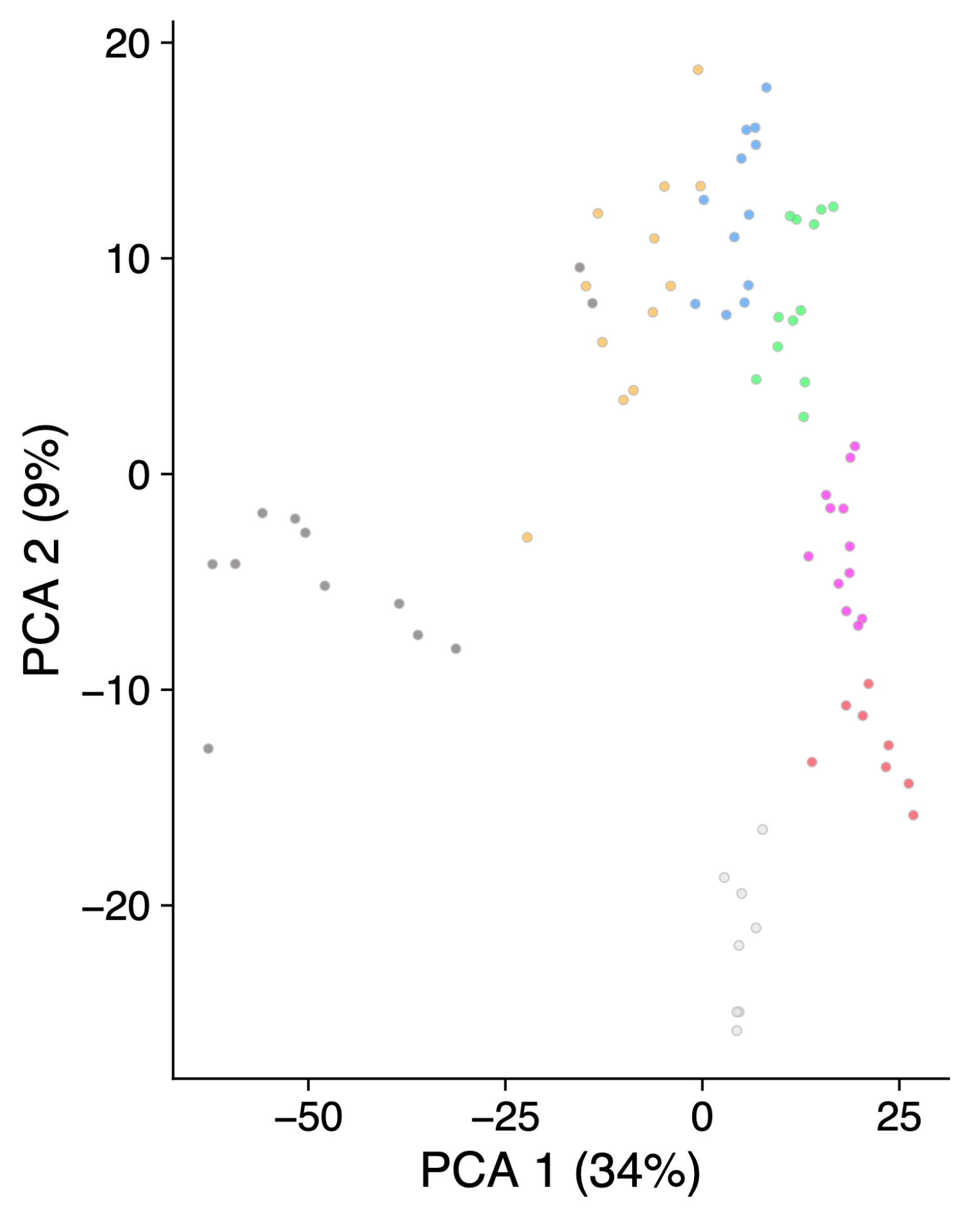
library('scater')
library('SummarizedExperiment')
plotReducedDim(
sce,
dimred = 'PCA',
colour_by = 'my_cluster_variable',
theme_size = 20
)
now looks like this:
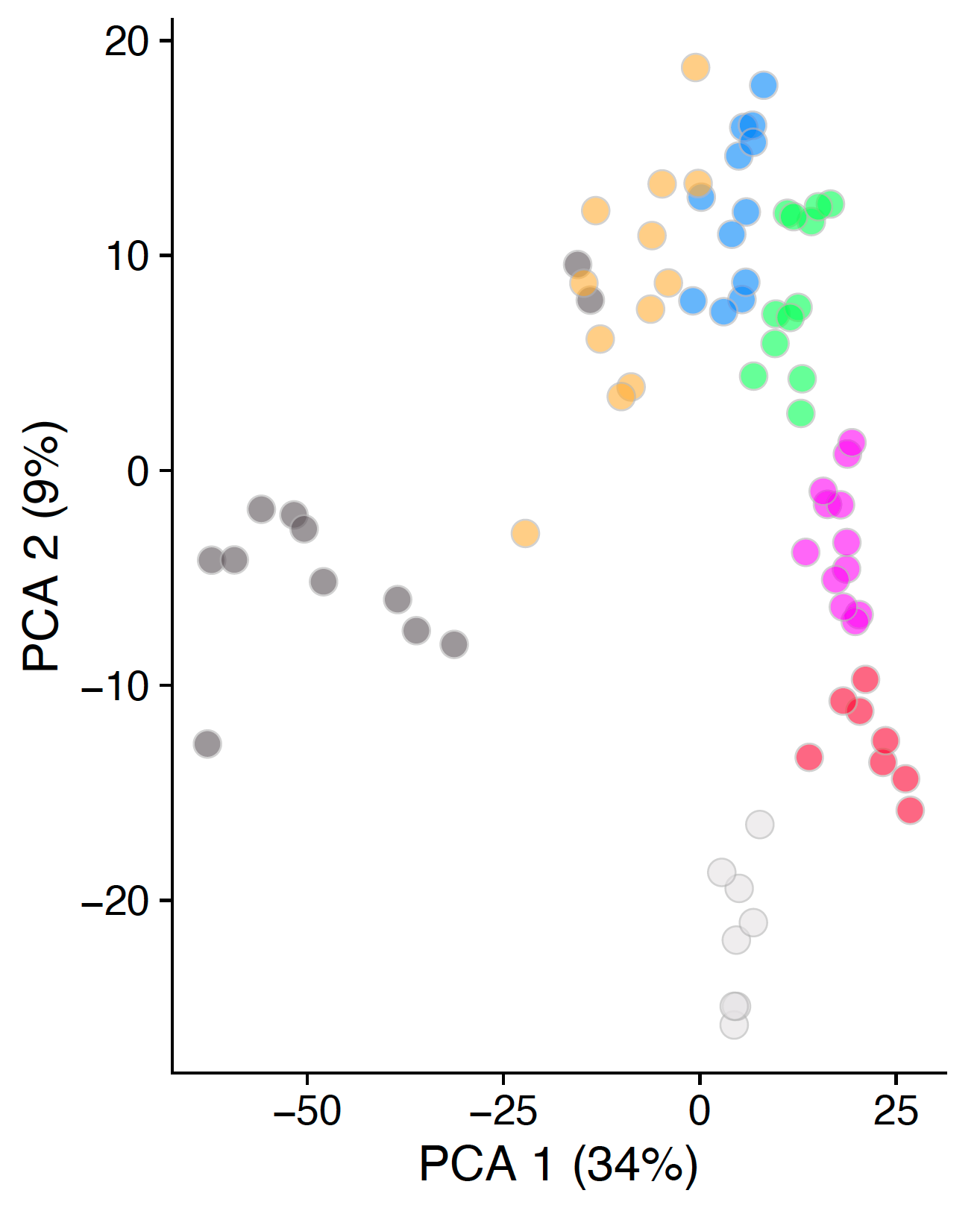
plotReducedDim(
sce,
dimred = 'PCA',
colour_by = 'my_cluster_variable',
theme_size = 20,
point_size = 5
)
Other examples involve different websites and showcase the diversity of questions we have as a team.
Conclusions
I hope that you like this idea and try it out yourselves. Maybe some of the lessons you learn trying it out could be useful to us as well. Ultimately, the information stored there could be useful for new team members as well as for current members since the spreadsheet becomes like an informal FAQ or team wiki.
I was strongly encouraged by the feedback two members gave me individually after our trial session. Maybe this is not for everyone as we realized that it’s quite hard to be anonymous while participating 7. This idea could evolve into something else, but at least for today, I’m happy with the amount of people that bought into the trial and participated in it. We’ll see what happens next.
Acknowledgments
This blog post was made possible thanks to:
- BiocStyle (Oleś, 2023)
- blogdown (Xie, Hill, and Thomas, 2017)
- ggplot2 (Wickham, 2016)
- knitcitations (Boettiger, 2021)
- scater (McCarthy, Campbell, Lun, and Willis, 2017)
- sessioninfo (Wickham, Chang, Flight, Müller et al., 2021)
- SummarizedExperiment (Morgan, Obenchain, Hester, and Pagès, 2023)
I would also like to acknowledge the general inspiration I’ve gotten from Alison Hill’s educational work. Once the rstudio::conf(2020) videos are available, check the work her intern Desirée de Leon showcased which is related to the following tweet.
Ever seen a giraffe that can fit in a teacup? Time to share the first draft of the R and stats project that Hasse Wallum and I have been working on for some time. More to come! #rstats 🦒 https://t.co/SOlBzop8vz
— Desirée De Leon (@dcossyle) August 14, 2019
P.S. dinámica in Spanish is used for a set of exercises that have a specific purpose in mind. I now realize that dynamic doesn’t hold the same meaning. Oh well 🤷🏾
References
\[1\] C. Boettiger. knitcitations: Citations for ‘Knitr’ Markdown Files. R package version 1.0.12. 2021. URL: https://CRAN.R-project.org/package=knitcitations.
\[2\] D. J. McCarthy, K. R. Campbell, A. T. L. Lun, and Q. F. Willis. “Scater: pre-processing, quality control, normalisation and visualisation of single-cell RNA-seq data in R”. In: Bioinformatics 33 (8 2017), pp. 1179-1186. DOI: 10.1093/bioinformatics/btw777.
\[3\] M. Morgan, V. Obenchain, J. Hester, and H. Pagès. SummarizedExperiment: SummarizedExperiment container. R package version 1.30.2. 2023. DOI: 10.18129/B9.bioc.SummarizedExperiment. URL: https://bioconductor.org/packages/SummarizedExperiment.
\[4\] A. Oleś. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.28.0. 2023. DOI: 10.18129/B9.bioc.BiocStyle. URL: https://bioconductor.org/packages/BiocStyle.
\[5\] H. Wickham. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016. ISBN: 978-3-319-24277-4. URL: https://ggplot2.tidyverse.org.
\[6\] H. Wickham, W. Chang, R. Flight, K. Müller, et al. sessioninfo: R Session Information. R package version 1.2.2. 2021. URL: https://CRAN.R-project.org/package=sessioninfo.
\[7\] Y. Xie, A. P. Hill, and A. Thomas. blogdown: Creating Websites with R Markdown. Boca Raton, Florida: Chapman and Hall/CRC, 2017. ISBN: 978-0815363729. URL: https://bookdown.org/yihui/blogdown/.
Reproducibility
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.3.1 (2023-06-16)
## os macOS Ventura 13.4
## system aarch64, darwin20
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/New_York
## date 2023-07-11
## pandoc 3.1.5 @ /opt/homebrew/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
## beachmat 2.16.0 2023-04-25 [1] Bioconductor
## beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.3.0)
## bibtex 0.5.1 2023-01-26 [1] CRAN (R 4.3.0)
## Biobase * 2.60.0 2023-04-25 [1] Bioconductor
## BiocGenerics * 0.46.0 2023-04-25 [1] Bioconductor
## BiocManager 1.30.21 2023-06-10 [1] CRAN (R 4.3.0)
## BiocNeighbors 1.18.0 2023-04-25 [1] Bioconductor
## BiocParallel 1.34.2 2023-05-28 [1] Bioconductor
## BiocSingular 1.16.0 2023-04-25 [1] Bioconductor
## BiocStyle * 2.28.0 2023-04-25 [1] Bioconductor
## bitops 1.0-7 2021-04-24 [1] CRAN (R 4.3.0)
## blogdown 1.18 2023-06-19 [1] CRAN (R 4.3.0)
## bookdown 0.34 2023-05-09 [1] CRAN (R 4.3.0)
## bslib 0.5.0 2023-06-09 [1] CRAN (R 4.3.0)
## cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
## cli 3.6.1 2023-03-23 [1] CRAN (R 4.3.0)
## codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.1)
## colorout 1.2-2 2023-05-06 [1] Github (jalvesaq/colorout@79931fd)
## colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
## crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
## DelayedArray 0.26.3 2023-05-28 [1] Bioconductor
## DelayedMatrixStats 1.22.0 2023-05-08 [1] Bioconductor
## digest 0.6.31 2022-12-11 [1] CRAN (R 4.3.0)
## dplyr 1.1.2 2023-04-20 [1] CRAN (R 4.3.0)
## evaluate 0.21 2023-05-05 [1] CRAN (R 4.3.0)
## fansi 1.0.4 2023-01-22 [1] CRAN (R 4.3.0)
## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
## generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
## GenomeInfoDb * 1.36.0 2023-05-08 [1] Bioconductor
## GenomeInfoDbData 1.2.10 2023-05-06 [1] Bioconductor
## GenomicRanges * 1.52.0 2023-04-25 [1] Bioconductor
## ggbeeswarm 0.7.2 2023-04-29 [1] CRAN (R 4.3.0)
## ggplot2 * 3.4.2 2023-04-03 [1] CRAN (R 4.3.0)
## ggrepel 0.9.3 2023-02-03 [1] CRAN (R 4.3.0)
## glue 1.6.2 2022-02-24 [1] CRAN (R 4.3.0)
## gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
## gtable 0.3.3 2023-03-21 [1] CRAN (R 4.3.0)
## htmltools 0.5.5 2023-03-23 [1] CRAN (R 4.3.0)
## httr 1.4.6 2023-05-08 [1] CRAN (R 4.3.0)
## IRanges * 2.34.0 2023-05-08 [1] Bioconductor
## irlba 2.3.5.1 2022-10-03 [1] CRAN (R 4.3.0)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
## jsonlite 1.8.7 2023-06-29 [1] CRAN (R 4.3.0)
## knitcitations * 1.0.12 2021-01-10 [1] CRAN (R 4.3.0)
## knitr 1.43 2023-05-25 [1] CRAN (R 4.3.0)
## lattice 0.21-8 2023-04-05 [1] CRAN (R 4.3.1)
## lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.3.0)
## lubridate 1.9.2 2023-02-10 [1] CRAN (R 4.3.0)
## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
## Matrix 1.5-4.1 2023-05-18 [1] CRAN (R 4.3.1)
## MatrixGenerics * 1.12.0 2023-05-08 [1] Bioconductor
## matrixStats * 1.0.0 2023-06-02 [1] CRAN (R 4.3.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
## plyr 1.8.8 2022-11-11 [1] CRAN (R 4.3.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
## Rcpp 1.0.10 2023-01-22 [1] CRAN (R 4.3.0)
## RCurl 1.98-1.12 2023-03-27 [1] CRAN (R 4.3.0)
## RefManageR 1.4.0 2022-09-30 [1] CRAN (R 4.3.0)
## rlang 1.1.1 2023-04-28 [1] CRAN (R 4.3.0)
## rmarkdown 2.23 2023-07-01 [1] CRAN (R 4.3.0)
## rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.3.0)
## rsvd 1.0.5 2021-04-16 [1] CRAN (R 4.3.0)
## S4Arrays 1.0.4 2023-05-14 [1] Bioconductor
## S4Vectors * 0.38.1 2023-05-02 [1] Bioconductor
## sass 0.4.6.9000 2023-05-06 [1] Github (rstudio/sass@f248fe5)
## ScaledMatrix 1.8.1 2023-05-03 [1] Bioconductor
## scales 1.2.1 2022-08-20 [1] CRAN (R 4.3.0)
## scater * 1.28.0 2023-04-25 [1] Bioconductor
## scuttle * 1.10.1 2023-05-02 [1] Bioconductor
## sessioninfo * 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
## SingleCellExperiment * 1.22.0 2023-04-25 [1] Bioconductor
## sparseMatrixStats 1.12.0 2023-05-08 [1] Bioconductor
## stringi 1.7.12 2023-01-11 [1] CRAN (R 4.3.0)
## stringr 1.5.0 2022-12-02 [1] CRAN (R 4.3.0)
## SummarizedExperiment * 1.30.2 2023-06-11 [1] Bioconductor
## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
## tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
## timechange 0.2.0 2023-01-11 [1] CRAN (R 4.3.0)
## utf8 1.2.3 2023-01-31 [1] CRAN (R 4.3.0)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.3.0)
## vipor 0.4.5 2017-03-22 [1] CRAN (R 4.3.0)
## viridis 0.6.3 2023-05-03 [1] CRAN (R 4.3.0)
## viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.0)
## withr 2.5.0 2022-03-03 [1] CRAN (R 4.3.0)
## xfun 0.39 2023-04-20 [1] CRAN (R 4.3.0)
## xml2 1.3.4 2023-04-27 [1] CRAN (R 4.3.0)
## XVector 0.40.0 2023-04-25 [1] Bioconductor
## yaml 2.3.7 2023-01-23 [1] CRAN (R 4.3.0)
## zlibbioc 1.46.0 2023-04-25 [1] Bioconductor
##
## [1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
-
So you can link to specific lines and see things you changed through time that might be the source of the problem are among the main reasons why you should try to version control your code. ↩︎
-
Potentially to a more interesting problem than the one you are stuck currently at. ↩︎
-
For example, some use a particular project Slack channel, others the lab one, others through direct messages. ↩︎
-
If you say that something is obvious or easy, you are telling the other person that it should be easy for them too, but we know that it isn’t the case and that’s why they are asking a question. ↩︎
-
If we want to incorporate this exercise into our weekly meetings (maybe once a month), we need to make sure that our team meeting will finish on time. ↩︎
-
The use of keywords can dramatically affect a search results, and this information is useful to convey among ourselves so we can learn to search for help more effectively. ↩︎
-
Basically, you can only be anonymous for those not in the room at the time the question was discussed. ↩︎