Diving together into the unknown world of spatial transcriptomics
Yesterday was an extremely exciting day for me and my colleagues. We finished a project we had been working on and shared it with the world. Meaning, it’s done and we can relax for a little bit while we wait for feedback from our peers.
But this was not any project, at least not for me. Why do you ask? In general terms, it involved an analysis that you could not search on Google and find the answer for. That is, it involved diving into the unknown!
The unknown is scary and as the lyrics say:
I’ve had my adventure, I don’t need something new I’m afraid of what I’m risking if I follow you Into the unknown
All of us have been building our careers with other types of data and/or experiments, and taking on a new type of data knowing we had an early access advantage over others was quite the challenge. I don’t know about my co-authors, but maybe some of them shared thoughts like mine that were along the lines: can I do this? can I make it work? do my analysis choices make sense? what will experts think of doing once they have access to this data? All while racing against time, even if it was just an illusion in our minds.
But it’s not my first adventure and I’ve picked up skills and confidence along the way. In particular, I’ve written Bioconductor R packages, dealt with pkgdown/travis issues like #1206, made shiny web applications, analyzed large RNA-seq data, written papers using GoogleDocs, gotten better at asking for help, among other skills.
It's Thurs but here's the "more" I promised https://t.co/4gahvWDbWE @travisci #pkgdown
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) February 28, 2020
+my fam of #rstats issues today https://t.co/iGFfHLtasM https://t.co/M8DZEHryJZ https://t.co/FU8qq3f5p6
I'm the pest that visits your GH issues here & there😅Nah, I put lots of 👨🏾💻into them!😊 https://t.co/1G0nd5ZwUJ
I’ve also gotten more comfortable with the idea that I can’t do it all. Others will shortly develop new methods for this type of data, or proper infrastructure to handle this data, or faster visualizations, and so goes on the list. But I’m proud and really happy to say that we built quite the robust prototype. Plus maybe we’ll be involved in shaping this future.
And you noticed that I mentioned we. That’s because I have been learning over the years how to foster collaborations. This particular project involved working with two other members of my workplace who are awesome and that I didn’t know that well. It also involved a new collaboration with someone I’ve known for a while now (we initially met through Twitter in 2014) but hadn’t had the chance to work with. Thus we dove into the unknown together 👩🚀🧑🚀.
I feel like we complemented each other quite well and all I can confidently say that our new adventure so far has been very stimulating, even it cost me some sleep.
Woke up at 3:30 am dreaming about code 👨🏻💻 😴 and on an off till 5:30. Gave up and went to work early... hopefully I can remember the dreams 😅#CodeWhileDreaming #RestFuelsIdeas pic.twitter.com/h0mYHAaL5W
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) February 19, 2020
Spatial transcriptomics
So, where does spatial transcriptomics come into play and what does it mean? I work with gene activity data which we formally refer to as gene expression 🧬. That is, we measure 🔍🧮 the activity levels of genes for a particular biological condition or tissue sample. For several years now (about since 2007-2009) we have been able to measure many genes from a tissue sample, called bulk RNA-sequencing and abbreviated as RNA-seq.
That’s great! But biology is complicated and a single tissue sample is composed of multiple cells of various types. For example, in the brain there are cells that send signals around (neurons) and others that give structure to the brain. That is why technologies for measuring the gene expression at the single cell level were developed, abbreviated as scRNA-seq. scRNA-seq has been used widely to study mouse brains to live tissue samples.
In recent years I’ve been working with data from the human brain 🧠. The Lieber Institute for Brain Development has about two thousand brain samples. To preserve them for years to come, the brains are frozen 🥶. Cells are a bit fragile and freezing them breaks them. This fact has made it challenging to study data from frozen human brains. Several of my colleagues work on adapting research protocols to handle frozen human brain tissue. The research field overall has been able to generate single nucleus RNA sequencing (snRNA-seq) data and we are all generating some more.
snRNA-seq and scRNA-seq are great because you can measure what genes (pieces of the cell) are active, classify them into groups, and use prior knowledge to label these groups. However, you lose information about what part of the tissue they come from. That’s where technologies for spatial transcriptomics, that is, measuring gene expression 🧬 as close a possible to the single cell level yet retaining spatial coordinates are being actively developed. Thus, you end up with two main sources of data: the gene expression measurements but also images from the tissue (histology information). My coworkers anticipated what could these technologies be used for and what type of research questions they help us answer.
oh hey, that's us! stay tuned for a biorxiv preprint! @LieberInstitute @martinowk @kr_maynard @fellgernon @stephaniehicks @lmwebr https://t.co/uYpwQu52pS
— Andrew Jaffe (@andrewejaffe) February 24, 2020
Our project’s history
My coworkers got early access to a specific new type of spatial transcriptomics technology called Visium from the 10x Genomics company and started piloting it on human brain tissue. They recruited me to their project in early November 2019 (11th) and I recruited more colleagues in early December (4th). Today on February 28th 2020 we made public our research advances, code, and software we built for this project.
Given that we have many potential websites others can find us through, we decided to unify as much as possible the documentation even if that meant repeating it. The basic start of our documentation is included further below.
spatialLIBD

Welcome to the spatialLIBD project! It is composed of:
- a shiny web application that we are hosting at spatial.libd.org/spatialLIBD/ that can handle a limited set of concurrent users,
- a Bioconductor package at bioconductor.org/packages/spatialLIBD (or from here) that lets you analyze the data and run a local version of our web application (with our data or yours),
- and a research article with the scientific knowledge we drew from this dataset. The analysis code for our project is available here and the high quality figures for the manuscript are available through Figshare.
The web application allows you to browse the LIBD human dorsolateral pre-frontal cortex (DLPFC) spatial transcriptomics data generated with the 10x Genomics Visium platform. Through the R/Bioconductor package you can also download the data as well as visualize your own datasets using this web application. Please check the manuscript or bioRxiv pre-print for more details about this project.
If you tweet about this website, the data or the R package please use the #spatialLIBD hashtag. You can find previous tweets that way as shown here. Thank you! Tweet #spatialLIBD
Study design
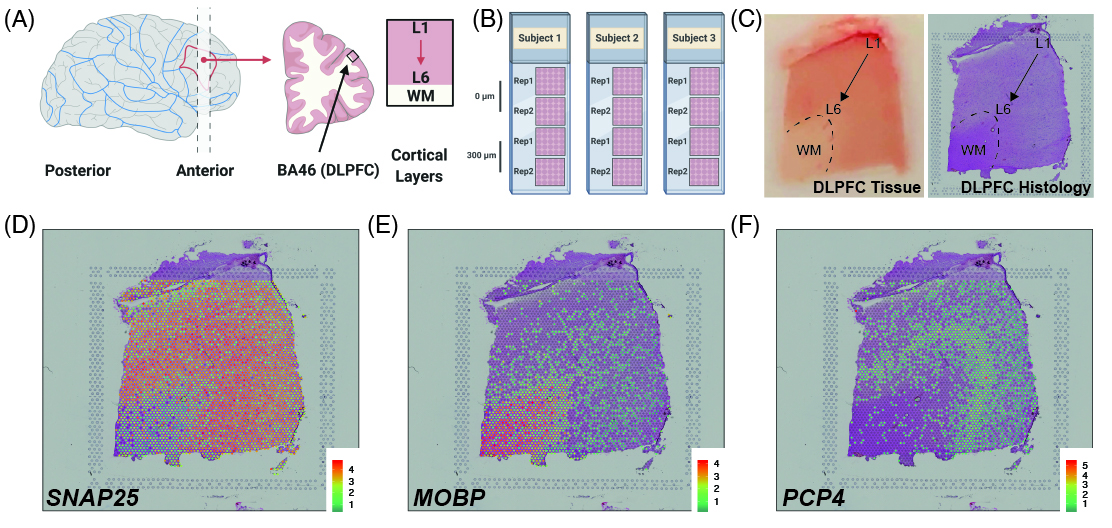
As a quick overview, the data presented here is from portion of the DLPFC that spans six neuronal layers plus white matter (A) for a total of three subjects with two pairs of spatially adjacent replicates (B). Each dissection of DLPFC was designed to span all six layers plus white matter (C). Using this web application you can explore the expression of known genes such as SNAP25 (D, a neuronal gene), MOBP (E, an oligodendrocyte gene), and known layer markers from mouse studies such as PCP4 (F, a known layer 5 marker gene).
Shiny website mirrors
- Main shiny application website (note that the link must have a trailing slash
/for it to work) - Shinyapps This version has less RAM memory but is typically deployed using the latest version of
spatialLIBD.
R/Bioconductor package
The spatialLIBD package contains functions for:
- Accessing the spatial transcriptomics data from the LIBD Human Pilot project (code on GitHub) generated with the Visium platform from 10x Genomics. The data is retrieved from Bioconductor’s
ExperimentHub. - Visualizing the spot-level spatial gene expression data and clusters.
- Inspecting the data interactively either on your computer or through spatial.libd.org/spatialLIBD/.
For more details, please check the documentation website or the Bioconductor package landing page here.
Installation instructions
Get the latest stable R release from CRAN. Then install spatialLIBD from Bioconductor using the following code:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("spatialLIBD")
Access the data
Through the spatialLIBD package you can access the processed data in it’s final R format. However, we also provide a table of links so you can download the raw data we received from 10x Genomics.
Processed data
Using spatialLIBD you can access the Human DLPFC spatial transcriptomics data from the 10x Genomics Visium platform. For example, this is the code you can use to access the layer-level data. For more details, check the help file for fetch_data().
## Load the package
library('spatialLIBD')
## Download the spot-level data
spe <- fetch_data(type = 'spe')
## This is a SpatialExperiment object
spe
## class: SpatialExperiment
## dim: 33538 47681
## metadata(0):
## assays(2): counts logcounts
## rownames(33538): ENSG00000243485 ENSG00000237613 ... ENSG00000277475
## ENSG00000268674
## rowData names(9): source type ... gene_search is_top_hvg
## colnames(47681): AAACAACGAATAGTTC-1 AAACAAGTATCTCCCA-1 ...
## TTGTTTCCATACAACT-1 TTGTTTGTGTAAATTC-1
## colData names(69): sample_id Cluster ... array_row array_col
## reducedDimNames(6): PCA TSNE_perplexity50 ... TSNE_perplexity80
## UMAP_neighbors15
## mainExpName: NULL
## altExpNames(0):
## spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
## imgData names(4): sample_id image_id data scaleFactor
## Note the memory size
lobstr::obj_size(spe)
## 2.04 GB
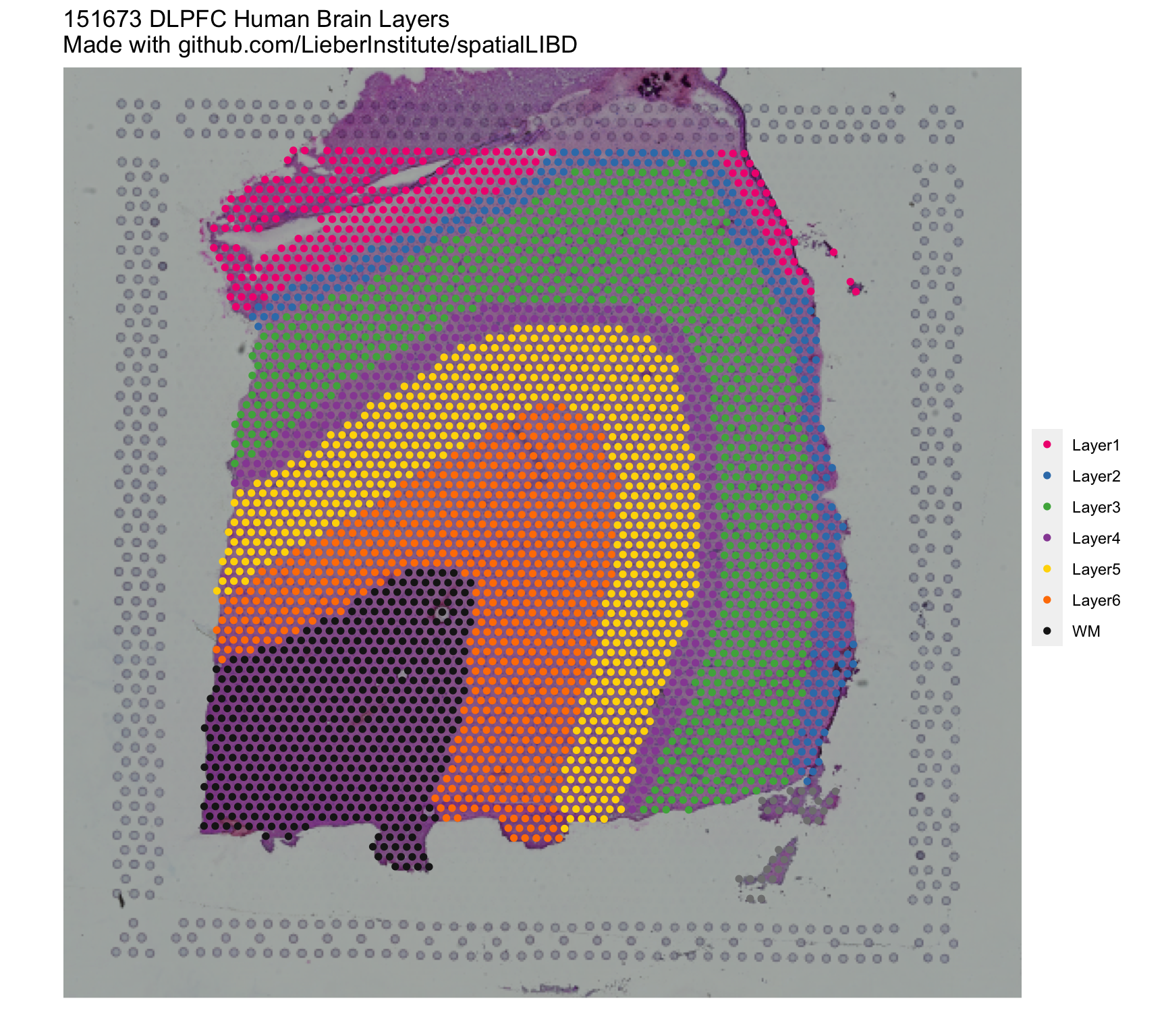
## Remake the logo image with histology information
vis_clus(
spe = spe,
clustervar = 'layer_guess_reordered',
sampleid = '151673',
colors = libd_layer_colors,
... = ' DLPFC Human Brain Layers\nMade with github.com/LieberInstitute/spatialLIBD'
)
Citation
Below is the citation output from using citation('spatialLIBD') in R. Please
run this yourself to check for any updates on how to cite spatialLIBD.
citation('spatialLIBD')
## To cite package 'spatialLIBD' in publications use:
##
## Pardo B, Spangler A, Weber LM, Hicks SC, Jaffe AE, Martinowich K,
## Maynard KR, Collado-Torres L (2022). "spatialLIBD: an R/Bioconductor
## package to visualize spatially-resolved transcriptomics data." _BMC
## Genomics_. doi:10.1186/s12864-022-08601-w
## <https://doi.org/10.1186/s12864-022-08601-w>,
## <https://doi.org/10.1186/s12864-022-08601-w>.
##
## Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK,
## Williams SR, II JLC, Tran MN, Besich Z, Tippani M, Chew J, Yin Y,
## Kleinman JE, Hyde TM, Rao N, Hicks SC, Martinowich K, Jaffe AE
## (2021). "Transcriptome-scale spatial gene expression in the human
## dorsolateral prefrontal cortex." _Nature Neuroscience_.
## doi:10.1038/s41593-020-00787-0
## <https://doi.org/10.1038/s41593-020-00787-0>,
## <https://www.nature.com/articles/s41593-020-00787-0>.
##
## Huuki-Myers LA, Spangler A, Eagles NJ, Montgomergy KD, Kwon SH, Guo
## B, Grant-Peters M, Divecha HR, Tippani M, Sriworarat C, Nguyen AB,
## Ravichandran P, Tran MN, Seyedian A, Consortium P, Hyde TM, Kleinman
## JE, Battle A, Page SC, Ryten M, Hicks SC, Martinowich K,
## Collado-Torres L, Maynard KR (2023). "Integrated single cell and
## unsupervised spatial transcriptomic analysis defines molecular
## anatomy of the human dorsolateral prefrontal cortex." _bioRxiv_.
## doi:10.1101/2023.02.15.528722
## <https://doi.org/10.1101/2023.02.15.528722>,
## <https://www.biorxiv.org/content/10.1101/2023.02.15.528722v1>.
##
## Kwon SH, Parthiban S, Tippani M, Divecha HR, Eagles NJ, Lobana JS,
## Williams SR, Mark M, Bharadwaj RA, Kleinman JE, Hyde TM, Page SC,
## Hicks SC, Martinowich K, Maynard KR, Collado-Torres L (2023).
## "Influence of Alzheimer’s disease related neuropathology on local
## microenvironment gene expression in the human inferior temporal
## cortex." _bioRxiv_. doi:10.1101/2023.04.20.537710
## <https://doi.org/10.1101/2023.04.20.537710>,
## <https://www.biorxiv.org/content/10.1101/2023.04.20.537710v1>.
##
## To see these entries in BibTeX format, use 'print(<citation>,
## bibtex=TRUE)', 'toBibtex(.)', or set
## 'options(citation.bibtex.max=999)'.
Please note that the spatialLIBD was only made possible thanks to many other R and bioinformatics software authors. We have cited their work either in the pre-print or the vignette of the R package.
Closing remarks
Overall, this project has everything that I like: R code, a Bioconductor package, challenging and interest biological data, excellent collaborator team, open communication, and so on.
Now, these are early days of the 10x Genomics Visium platform and there’s much we and others want to learn. So if you have the chance to hear anyone in our team talk more in detail about the project or you simply want to chat with them, here are some opportunities for you to do so as we’d love to collaborate with you or even hire you. Check Stephanie’s tweet and the LIBD career website for more details or simply get in touch with us.
- Kristen R Maynard and me will present a The Scientist webinar on March 19th
- Keri Martinowich will be at CVCSN 2020 March 26-27th
- I’ll present a seminar at LIIGH-UNAM on March 30th
- Kristen R Maynard will be at the 2020 Single Cell Symposium on April 20th
- Likely Andrew E Jaffe and others will be at The Biology of Genomes 2020 May 5-9th
- Stephanie Hicks will present at eRum 2020 May 27-30
- Likely some of us will attend BioC2020 July 29-31

Finally, here’s the pre-print twitter thread:
🔥off the press! 👀 our @biorxivpreprint on human 🧠brain @LieberInstitute spatial 🌌🔬transcriptomics data 🧬using Visium @10xGenomics🎉#spatialLIBD
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) February 29, 2020
🔍https://t.co/RTW0VscUKR
👩🏾💻https://t.co/bsg04XKONr
📚https://t.co/FJDOOzrAJ6
📦https://t.co/Au5jwADGhYhttps://t.co/PiWEDN9q2N pic.twitter.com/aWy0yLlR50
Thank you for getting this far!
References
Reproducibility
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.3.1 (2023-06-16)
## os macOS Ventura 13.4
## system aarch64, darwin20
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/New_York
## date 2023-07-11
## pandoc 3.1.5 @ /opt/homebrew/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## AnnotationDbi 1.62.1 2023-05-08 [1] Bioconductor
## AnnotationHub 3.8.0 2023-04-25 [1] Bioconductor
## attempt 0.3.1 2020-05-03 [1] CRAN (R 4.3.0)
## backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
## beachmat 2.16.0 2023-04-25 [1] Bioconductor
## beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.3.0)
## benchmarkme 1.0.8 2022-06-12 [1] CRAN (R 4.3.0)
## benchmarkmeData 1.0.4 2020-04-23 [1] CRAN (R 4.3.0)
## bibtex 0.5.1 2023-01-26 [1] CRAN (R 4.3.0)
## Biobase * 2.60.0 2023-04-25 [1] Bioconductor
## BiocFileCache 2.8.0 2023-04-25 [1] Bioconductor
## BiocGenerics * 0.46.0 2023-04-25 [1] Bioconductor
## BiocIO 1.10.0 2023-04-25 [1] Bioconductor
## BiocManager 1.30.21 2023-06-10 [1] CRAN (R 4.3.0)
## BiocNeighbors 1.18.0 2023-04-25 [1] Bioconductor
## BiocParallel 1.34.2 2023-05-28 [1] Bioconductor
## BiocSingular 1.16.0 2023-04-25 [1] Bioconductor
## BiocStyle * 2.28.0 2023-04-25 [1] Bioconductor
## BiocVersion 3.17.1 2022-12-20 [1] Bioconductor
## Biostrings 2.68.1 2023-05-16 [1] Bioconductor
## bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
## bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
## bitops 1.0-7 2021-04-24 [1] CRAN (R 4.3.0)
## blob 1.2.4 2023-03-17 [1] CRAN (R 4.3.0)
## blogdown 1.18 2023-06-19 [1] CRAN (R 4.3.0)
## bookdown 0.34 2023-05-09 [1] CRAN (R 4.3.0)
## bslib 0.5.0 2023-06-09 [1] CRAN (R 4.3.0)
## cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
## cli 3.6.1 2023-03-23 [1] CRAN (R 4.3.0)
## codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.1)
## colorout 1.2-2 2023-05-06 [1] Github (jalvesaq/colorout@79931fd)
## colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
## config 0.3.1 2020-12-17 [1] CRAN (R 4.3.0)
## cowplot 1.1.1 2020-12-30 [1] CRAN (R 4.3.0)
## crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
## curl 5.0.1 2023-06-07 [1] CRAN (R 4.3.0)
## data.table 1.14.8 2023-02-17 [1] CRAN (R 4.3.0)
## DBI 1.1.3 2022-06-18 [1] CRAN (R 4.3.0)
## dbplyr 2.3.2 2023-03-21 [1] CRAN (R 4.3.0)
## DelayedArray 0.26.3 2023-05-28 [1] Bioconductor
## DelayedMatrixStats 1.22.0 2023-05-08 [1] Bioconductor
## digest 0.6.31 2022-12-11 [1] CRAN (R 4.3.0)
## doParallel 1.0.17 2022-02-07 [1] CRAN (R 4.3.0)
## dotCall64 1.0-2 2022-10-03 [1] CRAN (R 4.3.0)
## dplyr 1.1.2 2023-04-20 [1] CRAN (R 4.3.0)
## dqrng 0.3.0 2021-05-01 [1] CRAN (R 4.3.0)
## DropletUtils 1.20.0 2023-05-08 [1] Bioconductor
## DT 0.28 2023-05-18 [1] CRAN (R 4.3.0)
## edgeR 3.42.4 2023-05-31 [1] Bioconductor
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
## evaluate 0.21 2023-05-05 [1] CRAN (R 4.3.0)
## ExperimentHub 2.8.0 2023-04-25 [1] Bioconductor
## fansi 1.0.4 2023-01-22 [1] CRAN (R 4.3.0)
## farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
## fields 14.1 2022-08-12 [1] CRAN (R 4.3.0)
## filelock 1.0.2 2018-10-05 [1] CRAN (R 4.3.0)
## foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
## generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
## GenomeInfoDb * 1.36.0 2023-05-08 [1] Bioconductor
## GenomeInfoDbData 1.2.10 2023-05-06 [1] Bioconductor
## GenomicAlignments 1.36.0 2023-04-25 [1] Bioconductor
## GenomicRanges * 1.52.0 2023-04-25 [1] Bioconductor
## ggbeeswarm 0.7.2 2023-04-29 [1] CRAN (R 4.3.0)
## ggplot2 3.4.2 2023-04-03 [1] CRAN (R 4.3.0)
## ggrepel 0.9.3 2023-02-03 [1] CRAN (R 4.3.0)
## glue 1.6.2 2022-02-24 [1] CRAN (R 4.3.0)
## golem 0.4.1 2023-06-05 [1] CRAN (R 4.3.0)
## gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
## gtable 0.3.3 2023-03-21 [1] CRAN (R 4.3.0)
## HDF5Array 1.28.1 2023-05-01 [1] Bioconductor
## highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
## htmltools 0.5.5 2023-03-23 [1] CRAN (R 4.3.0)
## htmlwidgets 1.6.2 2023-03-17 [1] CRAN (R 4.3.0)
## httpuv 1.6.11 2023-05-11 [1] CRAN (R 4.3.0)
## httr 1.4.6 2023-05-08 [1] CRAN (R 4.3.0)
## interactiveDisplayBase 1.38.0 2023-04-25 [1] Bioconductor
## IRanges * 2.34.0 2023-05-08 [1] Bioconductor
## irlba 2.3.5.1 2022-10-03 [1] CRAN (R 4.3.0)
## iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
## jsonlite 1.8.7 2023-06-29 [1] CRAN (R 4.3.0)
## KEGGREST 1.40.0 2023-04-25 [1] Bioconductor
## knitcitations * 1.0.12 2021-01-10 [1] CRAN (R 4.3.0)
## knitr 1.43 2023-05-25 [1] CRAN (R 4.3.0)
## labeling 0.4.2 2020-10-20 [1] CRAN (R 4.3.0)
## later 1.3.1 2023-05-02 [1] CRAN (R 4.3.0)
## lattice 0.21-8 2023-04-05 [1] CRAN (R 4.3.1)
## lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.3.0)
## lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.3.0)
## limma 3.56.2 2023-06-04 [1] Bioconductor
## lobstr 1.1.2 2022-06-22 [1] CRAN (R 4.3.0)
## locfit 1.5-9.8 2023-06-11 [1] CRAN (R 4.3.0)
## lubridate 1.9.2 2023-02-10 [1] CRAN (R 4.3.0)
## magick 2.7.4 2023-03-09 [1] CRAN (R 4.3.0)
## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
## maps 3.4.1 2022-10-30 [1] CRAN (R 4.3.0)
## Matrix 1.5-4.1 2023-05-18 [1] CRAN (R 4.3.1)
## MatrixGenerics * 1.12.0 2023-05-08 [1] Bioconductor
## matrixStats * 1.0.0 2023-06-02 [1] CRAN (R 4.3.0)
## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
## mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
## paletteer 1.5.0 2022-10-19 [1] CRAN (R 4.3.0)
## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
## plotly 4.10.2 2023-06-03 [1] CRAN (R 4.3.0)
## plyr 1.8.8 2022-11-11 [1] CRAN (R 4.3.0)
## png 0.1-8 2022-11-29 [1] CRAN (R 4.3.0)
## prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.3.0)
## promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.3.0)
## purrr 1.0.1 2023-01-10 [1] CRAN (R 4.3.0)
## R.methodsS3 1.8.2 2022-06-13 [1] CRAN (R 4.3.0)
## R.oo 1.25.0 2022-06-12 [1] CRAN (R 4.3.0)
## R.utils 2.12.2 2022-11-11 [1] CRAN (R 4.3.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
## rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.3.0)
## RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
## Rcpp 1.0.10 2023-01-22 [1] CRAN (R 4.3.0)
## RCurl 1.98-1.12 2023-03-27 [1] CRAN (R 4.3.0)
## RefManageR 1.4.0 2022-09-30 [1] CRAN (R 4.3.0)
## rematch2 2.1.2 2020-05-01 [1] CRAN (R 4.3.0)
## restfulr 0.0.15 2022-06-16 [1] CRAN (R 4.3.0)
## rhdf5 2.44.0 2023-04-25 [1] Bioconductor
## rhdf5filters 1.12.1 2023-04-30 [1] Bioconductor
## Rhdf5lib 1.22.0 2023-04-25 [1] Bioconductor
## rjson 0.2.21 2022-01-09 [1] CRAN (R 4.3.0)
## rlang 1.1.1 2023-04-28 [1] CRAN (R 4.3.0)
## rmarkdown 2.23 2023-07-01 [1] CRAN (R 4.3.0)
## Rsamtools 2.16.0 2023-04-25 [1] Bioconductor
## RSQLite 2.3.1 2023-04-03 [1] CRAN (R 4.3.0)
## rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.3.0)
## rsvd 1.0.5 2021-04-16 [1] CRAN (R 4.3.0)
## rtracklayer 1.60.0 2023-04-25 [1] Bioconductor
## S4Arrays 1.0.4 2023-05-14 [1] Bioconductor
## S4Vectors * 0.38.1 2023-05-02 [1] Bioconductor
## sass 0.4.6.9000 2023-05-06 [1] Github (rstudio/sass@f248fe5)
## ScaledMatrix 1.8.1 2023-05-03 [1] Bioconductor
## scales 1.2.1 2022-08-20 [1] CRAN (R 4.3.0)
## scater 1.28.0 2023-04-25 [1] Bioconductor
## scuttle 1.10.1 2023-05-02 [1] Bioconductor
## sessioninfo * 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
## shiny 1.7.4 2022-12-15 [1] CRAN (R 4.3.0)
## shinyWidgets 0.7.6 2023-01-08 [1] CRAN (R 4.3.0)
## SingleCellExperiment * 1.22.0 2023-04-25 [1] Bioconductor
## spam 2.9-1 2022-08-07 [1] CRAN (R 4.3.0)
## sparseMatrixStats 1.12.0 2023-05-08 [1] Bioconductor
## SpatialExperiment * 1.10.0 2023-04-25 [1] Bioconductor
## spatialLIBD * 1.13.4 2023-05-24 [1] Github (LieberInstitute/spatialLIBD@edc8b72)
## statmod 1.5.0 2023-01-06 [1] CRAN (R 4.3.0)
## stringi 1.7.12 2023-01-11 [1] CRAN (R 4.3.0)
## stringr 1.5.0 2022-12-02 [1] CRAN (R 4.3.0)
## SummarizedExperiment * 1.30.2 2023-06-11 [1] Bioconductor
## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
## tidyr 1.3.0 2023-01-24 [1] CRAN (R 4.3.0)
## tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
## timechange 0.2.0 2023-01-11 [1] CRAN (R 4.3.0)
## utf8 1.2.3 2023-01-31 [1] CRAN (R 4.3.0)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.3.0)
## vipor 0.4.5 2017-03-22 [1] CRAN (R 4.3.0)
## viridis 0.6.3 2023-05-03 [1] CRAN (R 4.3.0)
## viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.0)
## withr 2.5.0 2022-03-03 [1] CRAN (R 4.3.0)
## xfun 0.39 2023-04-20 [1] CRAN (R 4.3.0)
## XML 3.99-0.14 2023-03-19 [1] CRAN (R 4.3.0)
## xml2 1.3.4 2023-04-27 [1] CRAN (R 4.3.0)
## xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
## XVector 0.40.0 2023-04-25 [1] Bioconductor
## yaml 2.3.7 2023-01-23 [1] CRAN (R 4.3.0)
## zlibbioc 1.46.0 2023-04-25 [1] Bioconductor
##
## [1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
