remotes::install_github("jalvesaq/colorout")This lecture, as the rest of the course, is adapted from the version Stephanie C. Hicks designed and maintained in 2021 and 2022. Check the recent changes to this file through the GitHub history.
Pre-lecture materials
Read ahead
Before class, you can prepare by reading the following materials:
Acknowledgements
Material for this lecture was borrowed and adopted from
Learning objectives
At the end of this lesson you will:
- Discuss an overall approach to debugging code in R
- Recognize the three main indications of a problem/condition (
message,warning,error) and a fatal problem (error) - Understand the importance of reproducing the problem when debugging a function or piece of code
- Learn how to use interactive debugging tools
traceback,debug,recover,browser, andtracecan be used to find problematic code in functions
Debugging R Code
Overall approach
Finding the root cause of a problem is always challenging.
Most bugs are subtle and hard to find because if they were obvious, you would have avoided them in the first place.
A good strategy helps. Below I outline a five step process that I have found useful:
1. See it!
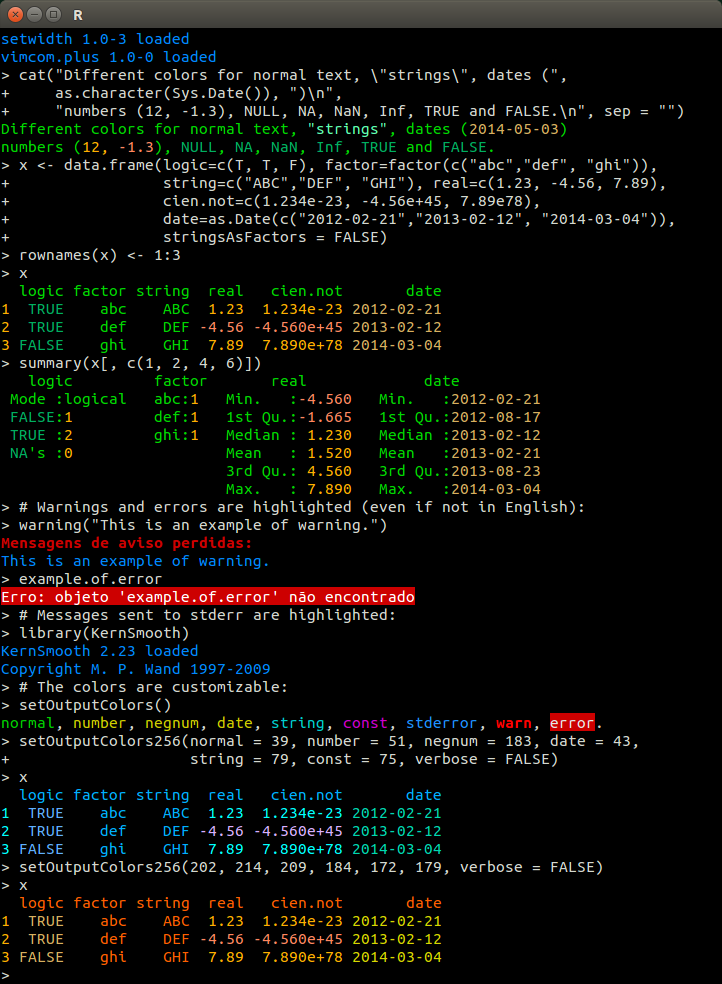
One of my favorite packages is colorout. It doesn’t work on winOS though 😢. Anyways, a very common mistake I see is that people don’t notice an earlier warning / error, which is actually more informative than the last error they get down the road. That can send people into a rabbit hole that doesn’t contain the relevant solution.
It works best if you load it automatically, which we can do by editing the configuration file called .Rprofile. It typically lives at ~/.Rprofile, but it’s best to access it with usethis::edit_r_profile().
## Open your .Rprofile file
usethis::edit_r_profile()
## Copy paste the following code taken from
## https://lcolladotor.github.io/bioc_team_ds/config-files.html#rprofile
## Change colors
# Source https://github.com/jalvesaq/colorout
if (Sys.getenv("TERM") %in% c("term", "xterm-256color", "cygwin", "screen")) {
if (!requireNamespace("colorout", quietly = TRUE) & .Platform$OS.type != "windows") {
cat('To install colorout use: remotes::install_github("jalvesaq/colorout")\n')
}
}Let’s give it a test.
I re-typed part of the code shown in the screenshot above. Compare how it looks in our RStudio window (if you are not on winOS) compared to how it looks on the HTML file for this lesson. Which one do you prefer?
require("colorout")
## From colorout's README documentation
x <- data.frame(
logic = c(TRUE, TRUE, FALSE),
factor = factor(c("abc", "def", "ghi")),
string = c("ABC", "DEF", "GHI"),
real = c(1.23, -4.56, 7.89),
cien.not = c(1.234e-23, -4.56 + 45, 7.89e78),
date = as.Date(c("2012-02-21", "2013-02-12", "2014-03-04"))
)
rownames(x) <- seq_len(3)
x logic factor string real cien.not date
1 TRUE abc ABC 1.23 1.234e-23 2012-02-21
2 TRUE def DEF -4.56 4.044e+01 2013-02-12
3 FALSE ghi GHI 7.89 7.890e+78 2014-03-04summary(x[, c(1, 2, 4, 6)]) logic factor real date
Mode :logical abc:1 Min. :-4.560 Min. :2012-02-21
FALSE:1 def:1 1st Qu.:-1.665 1st Qu.:2012-08-17
TRUE :2 ghi:1 Median : 1.230 Median :2013-02-12
Mean : 1.520 Mean :2013-02-21
3rd Qu.: 4.560 3rd Qu.:2013-08-23
Max. : 7.890 Max. :2014-03-04 warning("This is an example of a warning.")Warning: This is an example of a warning.example.of.errorError: object 'example.of.error' not foundlibrary("KernSmooth")KernSmooth 2.23 loaded
Copyright M. P. Wand 1997-2009colorout::setOutputColors()If you want to, use colorout::setOutputColors() to edit the colors.
2. Google!
Whenever you see an error message, start by googling it.
If you are lucky, you will discover that it’s a common error with a known solution.
When searching on Google or your preferred search browse, improve your chances of a good match by removing any variable names or values that are specific to your problem.
3. Make it repeatable
To find the root cause of an error, you are going to need to execute the code many times as you consider and reject hypotheses.
To make that iteration as quick possible, it’s worth some upfront investment to make the problem both easy and fast to reproduce.
Start by creating a reproducible example (reprex).
- This will help others help you, and often leads to a solution without asking others, because in the course of making the problem reproducible you often figure out the root cause.
Make the example minimal by removing code and simplifying data.
- As you do this, you may discover inputs that do not trigger the error. - Make note of them: they will be helpful when diagnosing the root cause.
Let’s try making a reprex using the reprex package (installed with the tidyverse)
library("reprex")Write a bit of code and copy it to the clipboard:
(y <- 1:4)
mean(y)Enter reprex() in the R Console. In RStudio, you’ll see a preview of your rendered reprex.
It is now ready and waiting on your clipboard, so you can paste it into, say, a GitHub issue.
One last step. Let’s go here and open up an issue on the course website or respond to our practice issue from earlier in the course:
We will paste in the code from our reprex.
Notice that if you are asking about code in this class, you might need to use the wd argument and set it to here::here(), that is reprex::reprex(wd = here::here()) as showcased at https://github.com/lcolladotor/jhustatcomputing/issues/5.
In RStudio, you can access reprex from the addins menu, which makes it even easier to point out your code and select the output format.
Several times in the process of building a reprex and then simplifying it as much as possible, I have found the source of my error.
Remember that we configured our RStudio’s global options such that:
Workspace: restore .RData into workspace at startup is turned off
Save workspace to .RData on exit is set to Never
Those two settings will save us many headaches!
4. Figure out where it is
It’s a great idea to adopt the scientific method here.
- Generate hypotheses
- Design experiments to test them
- Record your results
This may seem like a lot of work, but a systematic approach will end up saving you time.
Often a lot of time can be wasted relying on my intuition to solve a bug (“oh, it must be an off-by-one error, so I’ll just subtract 1 here”), when I would have been better off taking a systematic approach.
If this fails, you might need to ask help from someone else.
If you have followed the previous step, you will have a small example that is easy to share with others. That makes it much easier for other people to look at the problem, and more likely to help you find a solution.
5. Fix it and test it
Once you have found the bug, you need to figure out how to fix it and to check that the fix actually worked.
Again, it is very useful to have automated tests in place, which you can implement with testthat as showcased in the previous class.
- Not only does this help to ensure that you have actually fixed the bug, it also helps to ensure you have not introduced any new bugs in the process.
- In the absence of automated tests, make sure to carefully record the correct output, and check against the inputs that previously failed.
As a package developer, you might want to use simple reprex cases users report to you as sources for new unit tests in your package. Make sure to recognize the author(s) of the reprex and give them credit!
Something’s Wrong!
Once you have made the error repeatable, the next step is to figure out where it comes from.
R has a number of ways to indicate to you that something is not right.
There are different levels of indication that can be used, ranging from mere notification to fatal error. Executing any function in R may result in the following conditions.
message: A generic notification/diagnostic message produced by themessage()function; execution of the function continueswarning: An indication that something is wrong but not necessarily fatal; execution of the function continues. Warnings are generated by thewarning()functionerror: An indication that a fatal problem has occurred and execution of the function stops. Errors are produced by thestop()orstopifnot()functions.condition: A generic concept for indicating that something unexpected has occurred; programmers can create their own custom conditions if they want.
Here is an example of a warning that you might receive in the course of using R.
log(-1)Warning in log(-1): NaNs produced[1] NaNThis warning lets you know that taking the log of a negative number results in a NaN value because you can’t take the log of negative numbers.
Nevertheless, R doesn’t give an error, because it has a useful value that it can return, the NaN value.
The warning is just there to let you know that something unexpected happen.
Depending on what you are programming, you may have intentionally taken the log of a negative number in order to move on to another section of code.
Cases with NAs
Here is another function that is designed to print a message to the console depending on the nature of its input.
print_message <- function(x) {
if (x > 0) {
print("x is greater than zero")
} else {
print("x is less than or equal to zero")
}
invisible(x)
}This function is simple:
- It prints a message telling you whether
xis greater than zero or less than or equal to zero. - It also returns its input invisibly, which is a common practice with “print” functions.
Returning an object invisibly means that the return value does not get auto-printed when the function is called.
Take a hard look at the function above and see if you can identify any bugs or problems.
We can execute the function as follows.
print_message(1)[1] "x is greater than zero"The function seems to work fine at this point. No errors, warnings, or messages.
print_message(NA)Error in if (x > 0) {: missing value where TRUE/FALSE neededWhat happened?
- Well, the first thing the function does is test if
x > 0. - But you can’t do that test if
xis aNAorNaNvalue. - R doesn’t know what to do in this case so it stops with a fatal error.
We can fix this problem by anticipating the possibility of NA values and checking to see if the input is NA with the is.na() function.
print_message2 <- function(x) {
if (is.na(x)) {
print("x is a missing value!")
} else if (x > 0) {
print("x is greater than zero")
} else {
print("x is less than or equal to zero")
}
invisible(x)
}Now we can run the following.
print_message2(NA)[1] "x is a missing value!"And all is fine.
Cases with longer inputs than expected
Now what about the following situation.
x <- log(c(-1, 2))Warning in log(c(-1, 2)): NaNs producedprint_message2(x)Error in if (is.na(x)) {: the condition has length > 1Now what?? Why are we getting this warning?
The warning says “the condition has length > 1 and only the first element will be used”.
The problem here is that I passed print_message2() a vector x that was of length 2 rather then length 1.
Inside the body of print_message2() the expression is.na(x) returns a vector that is tested in the if statement.
However, if cannot take vector arguments, so you get a warning.
The fundamental problem here is that print_message2() is not vectorized.
We can solve this problem two ways.
- Simply not allow vector arguments.
- The other way is to vectorize the
print_message2()function to allow it to take vector arguments.
For the first way, we simply need to check the length of the input.
print_message3 <- function(x) {
if (length(x) > 1L) {
stop("'x' has length > 1")
}
if (is.na(x)) {
print("x is a missing value!")
} else if (x > 0) {
print("x is greater than zero")
} else {
print("x is less than or equal to zero")
}
invisible(x)
}Now when we pass print_message3() a vector, we should get an error.
print_message3(1:2)Error in print_message3(1:2): 'x' has length > 1Don’t show users the call to help them and you too!
I have learned that using that setting call. = FALSE when using stop() and/or warning() helps your users by providing them less information that could confuse them.
print_message3_no_call <- function(x) {
if (length(x) > 1L) {
stop("'x' has length > 1", call. = FALSE)
}
if (is.na(x)) {
print("x is a missing value!")
} else if (x > 0) {
print("x is greater than zero")
} else {
print("x is less than or equal to zero")
}
invisible(x)
}print_message3_no_call(99:100)Error: 'x' has length > 1print_message3(99:100)Error in print_message3(99:100): 'x' has length > 1If we compare the error output from print_message3() against print_message3_no_call() we can see that the second scenario doesn’t include information that only we (as the user) have. That’s information that users will copy-paste on their Google searches, which makes finding the right information much harder. As the person trying to help users, knowing how the users called our function is very likely not useful enough information. A reprex is 💯 * ♾️ much better!!!!!
Errors à la tidyverse
If you want to write error messages similar to those you are used to seeing with tidyverse packages, use rlang. Specifically, switch:
base::stop()withrlang::abort()base::warning()withrlang::warn()base::message()withrlang::inform()
print_message3_tidyverse <- function(x) {
if (length(x) > 1L) {
rlang::abort("'x' has length > 1")
}
if (is.na(x)) {
rlang::warn("x is a missing value!")
} else if (x > 0) {
rlang::inform("x is greater than zero")
} else {
rlang::inform("x is less than or equal to zero")
}
invisible(x)
}print_message3_tidyverse(99:100)Error in `print_message3_tidyverse()`:
! 'x' has length > 1print_message3_tidyverse(NA)Warning: x is a missing value!print_message3_tidyverse(1)x is greater than zeroprint_message3_tidyverse(-1)x is less than or equal to zeroNote how rlang by default doesn’t show the function call. The tidyverse style guide has a whole chapter on how to format error messages: https://style.tidyverse.org/error-messages.html. That is how all the R developers in the tidyverse team have been able to provide consistent looking messages to users of all these packages. They also use cli to make their error messages super pretty to read https://rlang.r-lib.org/reference/topic-condition-formatting.html.
In this next example, I:
use
cli::cli_abort()instead ofrlang::abort()define
lenso I can use it in a message with{len}use
{.code something}for inline-markup https://cli.r-lib.org/reference/inline-markup.htmluse several of the
clibullets, see https://cli.r-lib.org/reference/cli_bullets.html for the full listuse
praise::praise()to get some fun messages to praise our user and lift up their spirits =)- See also
praiseMXhttps://github.com/ComunidadBioInfo/praiseMX which CDSB students built in a course I co-instructed in 2019.
- See also
print_message3_cli <- function(x) {
if (length(x) > 1L) {
len <- length(x)
## Avoid the print() calls from
## https://github.com/ComunidadBioInfo/praiseMX/blob/master/R/praise_crear_emi.R
praise_mx_log <- capture.output({
praise_mx <- praiseMX:::praise_bien()
})
cli::cli_abort(
c(
"This function is not vectorized:",
"i" = "{.var x} has length {len}.",
"x" = "{.var x} must have length 1.",
">" = "Try using {.code purrr::map(x, print_message3_cli)} to loop your input {.var x} on this function.",
"v" = praise::praise(),
"v" = praise_mx
)
)
}
if (is.na(x)) {
rlang::warn("x is a missing value!")
} else if (x > 0) {
rlang::inform("x is greater than zero")
} else {
rlang::inform("x is less than or equal to zero")
}
invisible(x)
}set.seed(20230928)
print_message3_cli(-1:1)Error in `print_message3_cli()`:
! This function is not vectorized:
ℹ `x` has length 3.
✖ `x` must have length 1.
→ Try using `purrr::map(x, print_message3_cli)` to loop your input `x` on this
function.
✔ You are neat!
✔ ¡Ah chingá! Programas bien perrón.purrr::map(-1:1, print_message3_cli)x is less than or equal to zero
x is less than or equal to zero
x is greater than zero[[1]]
[1] -1
[[2]]
[1] 0
[[3]]
[1] 1Vectorizing
Vectorizing the function can be accomplished easily with the Vectorize() function.
print_message4 <- Vectorize(print_message2)
out <- print_message4(c(-1, 2))[1] "x is less than or equal to zero"
[1] "x is greater than zero"You can see now that the correct messages are printed without any warning or error.
I stored the return value of print_message4() in a separate R object called out.
This is because when I use the Vectorize() function it no longer preserves the invisibility of the return value.
The primary task of debugging any R code is correctly diagnosing what the problem is.
When diagnosing a problem with your code (or somebody else’s), it’s important first understand what you were expecting to occur.
Then you need to idenfity what did occur and how did it deviate from your expectations.
Some basic questions you need to ask are
- What was your input? How did you call the function?
- What were you expecting? Output, messages, other results?
- What did you get?
- How does what you get differ from what you were expecting?
- Were your expectations correct in the first place?
- Can you reproduce the problem (exactly)?
Being able to answer these questions is important not just for your own sake, but in situations where you may need to ask someone else for help with debugging the problem.
Seasoned programmers will be asking you these exact questions.
Think about the person who is going to receive your question. At https://lcolladotor.github.io/bioc_team_ds/how-to-ask-for-help.html I showcase some examples by Jim Hester and other information you might want to think about when asking for help.
Debugging Tools in R
R provides a number of tools to help you with debugging your code. The primary tools for debugging functions in R are
traceback(): prints out the function call stack after an error occurs; does nothing if there’s no error- The
tidyverseversion of this base R function isrlang::last_error()https://rlang.r-lib.org/reference/last_error.html. For base R, you will need to enable tracing withrlangwithrlang::global_entrace()https://rlang.r-lib.org/reference/global_entrace.html. It can provide much more user-friendly output.
- The
debug(): flags a function for “debug” mode which allows you to step through execution of a function one line at a timebrowser(): suspends the execution of a function wherever it is called and puts the function in debug modetrace(): allows you to insert debugging code into a function at specific placesrecover(): allows you to modify the error behavior so that you can browse the function call stack
These functions are interactive tools specifically designed to allow you to pick through a function. There is also the more blunt technique of inserting print() or cat() statements in the function.
Using traceback()
The traceback() function prints out the function call stack after an error has occurred.
The function call stack is the sequence of functions that was called before the error occurred.
For example, you may have a function a() which subsequently calls function b() which calls c() and then d().
If an error occurs, it may not be immediately clear in which function the error occurred.
The traceback() function shows you how many levels deep you were when the error occurred.
Let’s use the mean() function on a vector z that does not exist in our R environment
> mean(z)
Error in mean(z) : object 'z' not found
> traceback()
1: mean(z)Here, it’s clear that the error occurred inside the mean() function because the object z does not exist.
The traceback() function must be called immediately after an error occurs. Once another function is called, you lose the traceback.
Here is a slightly more complicated example using the lm() function for linear modeling.
> lm(y ~ x)
Error in eval(expr, envir, enclos) : object ’y’ not found
> traceback()
7: eval(expr, envir, enclos)
6: eval(predvars, data, env)
5: model.frame.default(formula = y ~ x, drop.unused.levels = TRUE)
4: model.frame(formula = y ~ x, drop.unused.levels = TRUE)
3: eval(expr, envir, enclos)
2: eval(mf, parent.frame())
1: lm(y ~ x)You can see now that the error did not get thrown until the 7th level of the function call stack, in which case the eval() function tried to evaluate the formula y ~ x and realized the object y did not exist.
With rlang, this is how it look:
lm(y ~ x)Error in eval(predvars, data, env): object 'y' not foundrlang::last_error()Error: Can't show last error because no error was recorded yetNote that we need to use rlang::global_entrace() first since lm() is a base R function. If we do so, then rlang::last_error() does work.
> rlang::global_entrace()
> lm(y ~ x)
Error:
! object 'y' not found
Run `rlang::last_trace()` to see where the error occurred.
> rlang::last_error()
<error/rlang_error>
Error:
! object 'y' not found
---
Backtrace:
▆
1. └─stats::lm(y ~ x)
2. └─base::eval(mf, parent.frame())
3. └─base::eval(mf, parent.frame())Looking at the traceback is useful for figuring out roughly where an error occurred but it’s not useful for more detailed debugging. For that you might turn to the debug() function.
Using debug()
Click here for how to use debug() with an interactive browser.
The debug() function initiates an interactive debugger (also known as the “browser” in R) for a function. With the debugger, you can step through an R function one expression at a time to pinpoint exactly where an error occurs.
The debug() function takes a function as its first argument. Here is an example of debugging the lm() function.
> debug(lm) ## Flag the 'lm()' function for interactive debugging
> lm(y ~ x)
debugging in: lm(y ~ x)
debug: {
ret.x <- x
ret.y <- y
cl <- match.call()
...
if (!qr)
z$qr <- NULL
z
}
Browse[2]>Now, every time you call the lm() function it will launch the interactive debugger. To turn this behavior off you need to call the undebug() function.
The debugger calls the browser at the very top level of the function body. From there you can step through each expression in the body. There are a few special commands you can call in the browser:
nexecutes the current expression and moves to the next expressionccontinues execution of the function and does not stop until either an error or the function exitsQquits the browser
Here’s an example of a browser session with the lm() function.
Browse[2]> n ## Evalute this expression and move to the next one
debug: ret.x <- x
Browse[2]> n
debug: ret.y <- y
Browse[2]> n
debug: cl <- match.call()
Browse[2]> n
debug: mf <- match.call(expand.dots = FALSE)
Browse[2]> n
debug: m <- match(c("formula", "data", "subset", "weights", "na.action",
"offset"), names(mf), 0L)While you are in the browser you can execute any other R function that might be available to you in a regular session. In particular, you can use ls() to see what is in your current environment (the function environment) and print() to print out the values of R objects in the function environment.
You can turn off interactive debugging with the undebug() function.
undebug(lm) ## Unflag the 'lm()' function for debuggingUsing recover()
Click here for how to use recover() with an interactive browser.
The recover() function can be used to modify the error behavior of R when an error occurs. Normally, when an error occurs in a function, R will print out an error message, exit out of the function, and return you to your workspace to await further commands.
With recover() you can tell R that when an error occurs, it should halt execution at the exact point at which the error occurred. That can give you the opportunity to poke around in the environment in which the error occurred. This can be useful to see if there are any R objects or data that have been corrupted or mistakenly modified.
> options(error = recover) ## Change default R error behavior
> read.csv("nosuchfile") ## This code doesn't work
Error in file(file, "rt") : cannot open the connection
In addition: Warning message:
In file(file, "rt") :
cannot open file ’nosuchfile’: No such file or directory
Enter a frame number, or 0 to exit
1: read.csv("nosuchfile")
2: read.table(file = file, header = header, sep = sep, quote = quote, dec =
3: file(file, "rt")
Selection:The recover() function will first print out the function call stack when an error occurrs. Then, you can choose to jump around the call stack and investigate the problem. When you choose a frame number, you will be put in the browser (just like the interactive debugger triggered with debug()) and will have the ability to poke around.
Summary
- There are three main indications of a problem/condition:
message,warning,error; only anerroris fatal- We saw how to trigger them with base R functions such as
stop(), andtidyversefunctionsrlang::abort()andcli::cli_abort().
- We saw how to trigger them with base R functions such as
- When analyzing a function with a problem, make sure you can reproduce the problem, clearly state your expectations and how the output differs from your expectation
- Interactive debugging tools
traceback,debug,recover,browser, andtracecan be used to find problematic code in functions - Debugging tools are not a substitute for thinking!
Post-lecture materials
Final Questions
Here are some post-lecture questions to help you think about the material discussed.
- Try using
traceback()to debug this piece of code:
f <- function(a) g(a)
g <- function(b) h(b)
h <- function(c) i(c)
i <- function(d) {
if (!is.numeric(d)) {
stop("`d` must be numeric", call. = FALSE)
}
d + 10
}
f("a")Error: `d` must be numericDescribe in words what is happening above?
Additional Resources
R session information
options(width = 120)
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
setting value
version R version 4.5.1 (2025-06-13)
os macOS Sequoia 15.6.1
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz America/New_York
date 2025-09-24
pandoc 3.7.0.2 @ /opt/homebrew/bin/ (via rmarkdown)
quarto 1.4.550 @ /Applications/quarto/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
cli 3.6.5 2025-04-23 [1] CRAN (R 4.5.0)
colorout * 1.3-2 2025-05-09 [1] Github (jalvesaq/colorout@572ab10)
digest 0.6.37 2024-08-19 [1] CRAN (R 4.5.0)
evaluate 1.0.5 2025-08-27 [1] CRAN (R 4.5.0)
fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.5.0)
fs 1.6.6 2025-04-12 [1] CRAN (R 4.5.0)
glue 1.8.0 2024-09-30 [1] CRAN (R 4.5.0)
htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.5.0)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.5.0)
jsonlite 2.0.0 2025-03-27 [1] CRAN (R 4.5.0)
KernSmooth * 2.23-26 2025-01-01 [1] CRAN (R 4.5.1)
knitr 1.50 2025-03-16 [1] CRAN (R 4.5.0)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.5.0)
magrittr 2.0.4 2025-09-12 [1] CRAN (R 4.5.0)
pillar 1.11.1 2025-09-17 [1] CRAN (R 4.5.0)
praise 1.0.0 2015-08-11 [1] CRAN (R 4.5.0)
praiseMX 0.0.0.9000 2025-09-24 [1] Github (ComunidadBioInfo/praiseMX@9d26399)
purrr 1.1.0 2025-07-10 [1] CRAN (R 4.5.0)
reprex * 2.1.1 2024-07-06 [1] CRAN (R 4.5.0)
rlang 1.1.6 2025-04-11 [1] CRAN (R 4.5.0)
rmarkdown 2.29 2024-11-04 [1] CRAN (R 4.5.0)
sessioninfo 1.2.3 2025-02-05 [1] CRAN (R 4.5.0)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.5.0)
withr 3.0.2 2024-10-28 [1] CRAN (R 4.5.0)
xfun 0.53 2025-08-19 [1] CRAN (R 4.5.0)
yaml 2.3.10 2024-07-26 [1] CRAN (R 4.5.0)
[1] /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────