Measurement, Summary, and Methodological Variation in RNA-sequencing in Statistical Analysis of Next Generation Sequencing Data
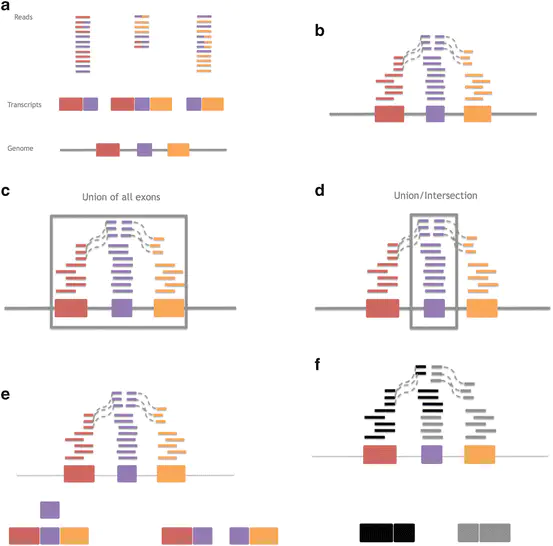 Image credit: Springer
Image credit: Springer
Abstract
There has been a major shift from microarrays to RNA-sequencing (RNA-seq) for measuring gene expression as the price per measurement between these technologies has become comparable. The advantages of RNA-seq are increased measurement flexibility to detect alternative transcription, allele specific transcription, or transcription outside of known coding regions. The price of this increased flexibility is: (a) an increase in raw data size and (b) more decisions that must be made by the data analyst. Here we provide a selective review and extension of our previous work in attempting to measure variability in results due to different choices about how to summarize and analyze RNA-sequencing data. We discuss a standard model for gene expression measurements that breaks variability down into variation due to technology, biology, and measurement error. Finally, wee show the importance of gene model selection, normalization, and choice for statistical model on the ultimate results of an RNA-sequencing experiment.