Integrated Transcriptomic and Proteomic Analysis of Primary Human Umbilical Vein Endothelial Cells
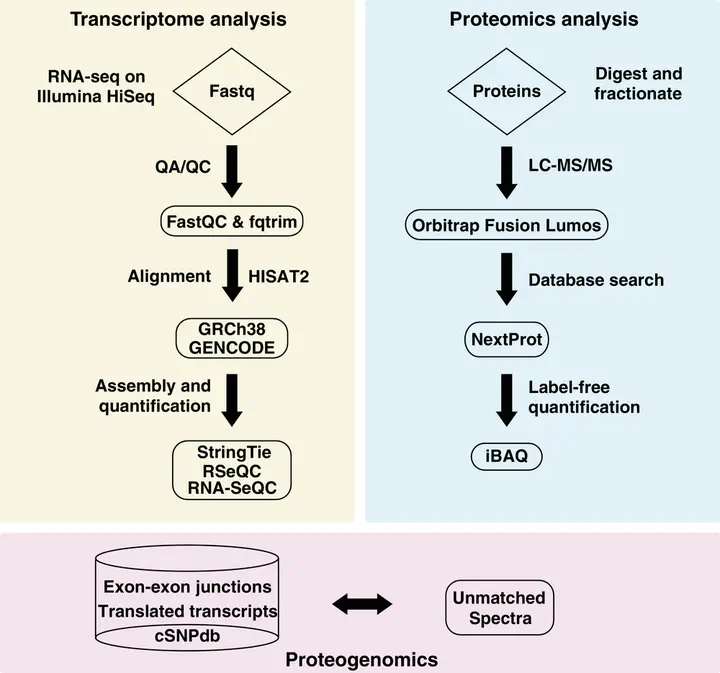 Image credit: Proteomics
Image credit: Proteomics
Abstract
Understanding the molecular profile of every human cell type is essential for understanding its role in normal physi-ology and disease. Technological advancements in DNA sequencing, mass spectrometry, and computational methodsallow us to carry out multiomics analyses although such approaches are not routine yet. Human umbilical vein endothe-lial cells (HUVECs) are a widely used model system to study pathological and physiological processes associated withthe cardiovascular system. In this study, next-generation sequencing and high-resolution mass spectrometry to profilethe transcriptome and proteome of primary HUVECs is employed. Analysis of 145 million paired-end reads from next-generation sequencing confirmed expression of 12 186 protein-coding genes (FPKM >= 0.1), 439 novel long non-codingRNAs, and revealed 6089 novel isoforms that were not annotated in GENCODE. Proteomics analysis identifies 6477proteins including confirmation ofN-termini for 1091 proteins, isoforms for 149 proteins, and 1034 phosphosites. Adatabase search to specifically identify other post-translational modifications provide evidence for a number of modifi-cation sites on 117 proteins which include ubiquitylation, lysine acetylation, and mono-, di- and tri-methylation events.Evidence for 11 ‘missing proteins’, which are proteins for which there was insufficient or no protein level evidence, isprovided. Peptides supporting missing protein and novel events are validated by comparison of MS/MS fragmentationpatterns with synthetic peptides. Finally, 245 variant peptides derived from 207 expressed proteins in addition to alternatetranslational start sites for seven proteins and evidence for novel proteoforms for five proteins resulting from alternativesplicing are identified. Overall, it is believed that the integrated approach employed in this study is widely applicable tostudy any primary cell type for deeper molecular characterization.
I'm glad to see published this project lead by Anil Madugundu in Akhilesh Pandey's labhttps://t.co/MF3PGL18et @proteomics
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) August 14, 2019
It was our first #recount2 collab w/ @Shannon_E_Ellis @chrisnwilks @BenLangmead where we brainstormed how to use the exon-exon junction data for proteomics pic.twitter.com/TI5PWq19Ok