 Image credit: Genome Research
Image credit: Genome Research
Abstract
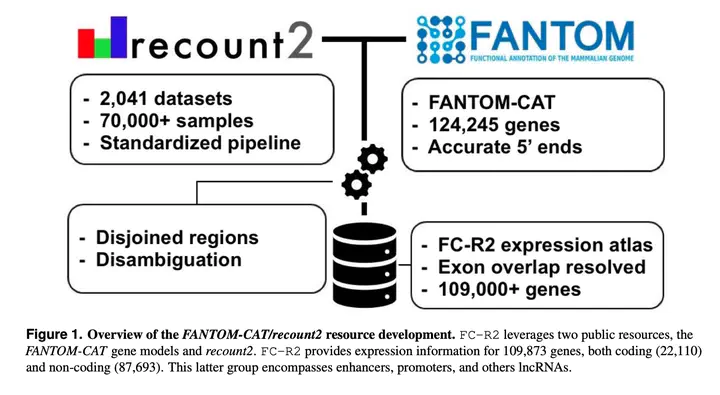
Long noncoding RNAs (lncRNAs) have emerged as key coordinators of biological and cellular processes. Characterizing lncRNA expression across cells and tissues is key to understanding their role in determining phenotypes including human diseases. We present here FC-R2, a comprehensive expression atlas across a broadly defined human transcriptome, inclusive of over 109,000 coding and noncoding genes, as described in the FANTOM CAGE-Associated Transcriptome (FANTOM-CAT) study. This atlas greatly extends the gene annotation used in the original recount2 resource. We demonstrate the utility of the FC-R2 atlas by reproducing key findings from published large studies and by generating new results across normal and diseased human samples. In particular, we (a) identify tissue-specific transcription profiles for distinct classes of coding and noncoding genes, (b) perform differential expression analysis across thirteen cancer types, identifying novel noncoding genes potentially involved in tumor pathogenesis and progression, and (c) confirm the prognostic value for several enhancers lncRNAs expression in cancer. Our resource is instrumental for the systematic molecular characterization of lncRNA by the FANTOM6 Consortium. In conclusion, comprised of over 70,000 samples, the FC-R2 atlas will empower other researchers to investigate functions and biological roles of both known coding genes and novel lncRNAs.
Congrats to Eddie-Luidy @eddieimada Diego @difersanchez & Luigi @marchionniLab et al for our work published in advance at @genomeresearch https://t.co/hIKaqs8XAm 🎉
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) February 21, 2020
It was powered by #recount2's BigWig files https://t.co/tK6iRfDhDr & @FANTOM_project's annotation #RNAseq #rstats pic.twitter.com/MTzZsftpyC