Programmatic access to bacterial regulatory networks with regutools
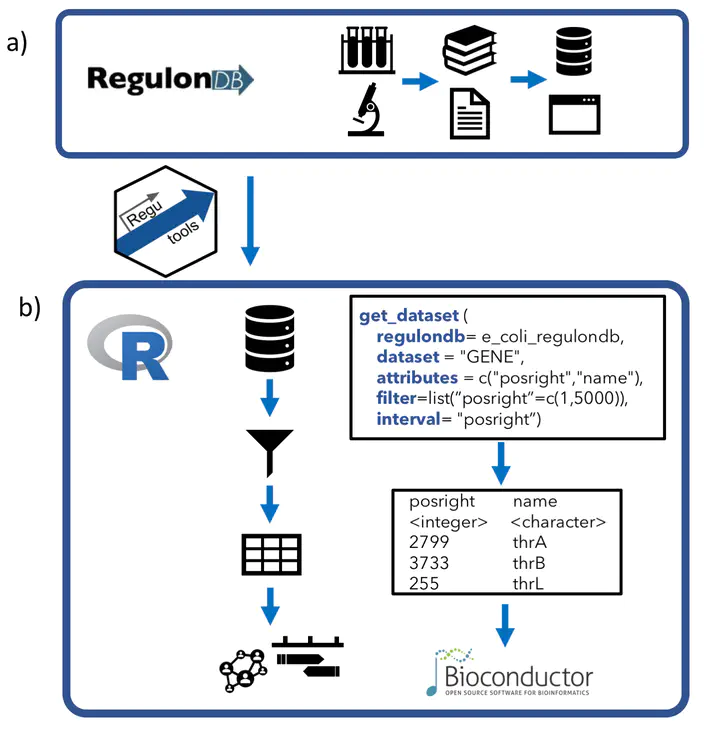 Image credit: Oxford Bioinformatics
Image credit: Oxford Bioinformatics
Abstract
RegulonDB has collected, harmonized and centralized data from hundreds of experiments for nearly two decades and is considered a point of reference for transcriptional regulation in Escherichia coli K12. Here, we present the regutools R package to facilitate programmatic access to RegulonDB data in computational biology. regutools gives researchers the possibility of writing reproducible workflows with automated queries to RegulonDB. The regutools package serves as a bridge between RegulonDB data and the Bioconductor ecosystem by reusing the data structures and statistical methods powered by other Bioconductor packages. We demonstrate the integration of regutools with Bioconductor by analyzing transcription factor DNA binding sites and transcriptional regulatory networks from RegulonDB. We anticipate that regutools will serve as a useful building block in our progress to further our understanding of gene regulatory networks.
The #regutools paper has been published by @OUPBioinfo. Thanks for your excellent work @BarjonCar, @josschavezf1, @EmilianoSotel10, @fellgernon, et al. First paper result of our efforts to increase the representation of #latam @Bioconductor developers. https://t.co/By1dEgxtGy https://t.co/WWsAYZ7KHM
— Alejandro Reyes (@areyesq) June 24, 2020