Data-driven identification of total RNA expression genes for estimation of RNA abundance in heterogeneous cell types highlighted in brain tissue
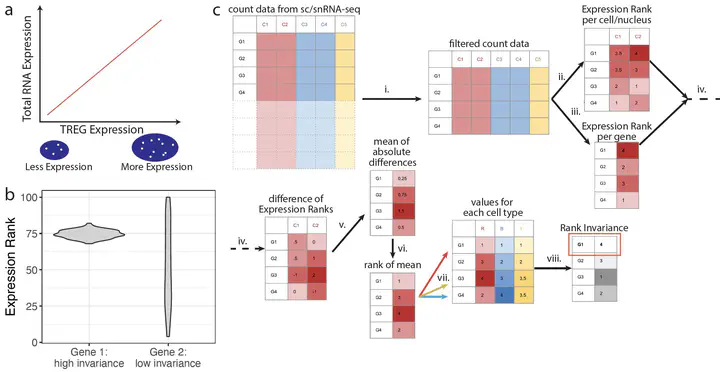 Image credit: bioRxiv
Image credit: bioRxiv
Abstract
We define and identify a new class of control genes for next-generation sequencing called total RNA expression genes (TREGs), which correlate with total RNA abundance in cell types of different sizes and transcriptional activity. We provide a data-driven method to identify TREGs from single-cell RNA sequencing data, allowing the estimation of total amount of RNA when restricted to quantifying a limited number of genes. We demonstrate our method in postmortem human brain using multiplex single-molecule fluorescent in situ hybridization and compare candidate TREGs against classic housekeeping genes. We identify AKT3 as a top TREG across five brain regions.
Excited to share my first 🥇 first-author manuscript: "Data-driven Identification of Total RNA Expression Genes (TREGs) for Estimation of RNA Abundance in Heterogeneous Cell Types" 👩🔬 @LieberInstitute #scitwitter
— Louise Huuki-Myers (@lahuuki) May 3, 2022
Now a @biorxivpreprint ! 🎉
📰 https://t.co/OJuAWndW51 pic.twitter.com/VZ8xzJpcyv
To end the week: CONGRATS @lahuuki for your first first-author @biorxivpreprint 📜! 🎉 The first of many I'm 💯 sure!#rstats @LieberInstitute
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) April 30, 2022
📦@Bioconductor https://t.co/5f79NtYyFu
💻@GitHub https://t.co/Hpg8rKLsst
📔#pkgdown https://t.co/6of1LCtxgC
👀https://t.co/J0uJA15liE pic.twitter.com/uUPZEUXns9