lute: estimating the cell composition of heterogeneous tissue with varying cell sizes using gene expression
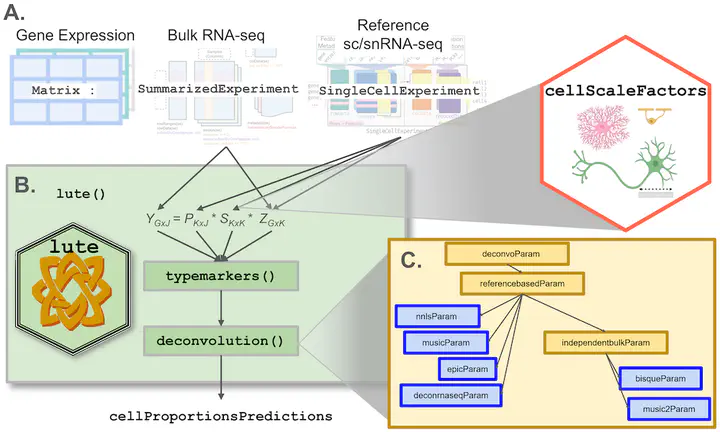 Image credit: bioRxiv
Image credit: bioRxiv
Abstract
Relative cell type fraction estimates in bulk RNA-sequencing data are important to control for cell composition differences across heterogenous tissue samples. Current computational tools estimate relative RNA abundances rather than cell type proportions in tissues with varying cell sizes, leading to biased estimates. We present lute, a computational tool to accurately deconvolute cell types with varying sizes. Our software wraps existing deconvolution algorithms in a standardized framework. Using simulated and real datasets, we demonstrate how lute adjusts for differences in cell sizes to improve the accuracy of cell composition. Software is available from https://bioconductor.org/packages/lute.
1/🎉Ecstatic the lute software preprint with my mentor @stephaniehicks deconvolving sn & bulk #rnaseq of human brain is now on #bioRxiv! Thankful to @jhubiostat,@LieberInstitute, and my brilliant colleagues @lahuuki @sanghokwon17 @lcolladotor @kr_maynard 🎉https://t.co/OZKDY9vECL
— Sean Maden (@MadenSean) April 22, 2024