Login to the cluster, request a node and change to your project directory in a single command
To be able to do RNA-seq research work in large multi-sample studies you have to be able to analyze large files and thus frequently use a powerful computing environment. In my case, this means that I have to login to a computing cluster frequently. This is a common task for other biostatisticians (like those that do brain imagining studies) and many other people. When I am working on a project, I generally have to login to the cluster and then change the directory to the location where I have my project files.
A local cluster is normally composed of a login machine (enigma2 in my case) from where you can request to work on a node. There are several options for controlling this process of requesting a node, and in our institution we use a Sun/Oracle Grid Engine. When a cluster has a lot of users, you want to dedicate the login machine as much as possible to handling login requests and assigning nodes for people to use. This means that you want to minimize doing any other kind of operations on the login machine, such as input/output operations.
Basic workflow
This means that the steps I normally follow before getting to work on my project are:
## Open the terminal
## Login to enigma
ssh username@enigma2.etc.edu
## Request a node to work on interactively
qrsh
## Change to the directory where I have my project files
cd projectDir
## Done!
For a long time I have used an alias for the login step. I have this alias in my local .bashrc file:
## In local .bashrc file
alias enigma="ssh username@enigma2.etc.edu"
Thus saving a tiny bit of typing:
## Open terminal
## Login to enigma
enigma
## Etc
qrsh
cd projectDir
When the projectDir gets complicated, I make an alias on my .bashrc file on the cluster. For example:
## In cluster .bashrc file
alias pdir="cd /very/complicated/path/to/projectDir/"
And finally, I can accomplish the setup task with minimal typing:
## Open terminal
enigma
qrsh
pdir
Using ssh config
This section was added after Kasper Hansen’s comment.
You can edit the ~.ssh/config file (check how to set it up, explained differently and the manual) to make things even better. This is how mine looks like:
Host enigma
User username
Hostname enigma2.etc.edu
ForwardX11 yes
I like the ssh -X (or -Y) option so I can later view plots in X11 when running R. That is why the ForwardX11 option is present.
Then you can use the following command to ssh into the cluster.
ssh enigma
Or if you prefer, simplify the bash alias to:
## In local .bashrc file
alias enigma="ssh enigma"
The sections below have been edited to assume that you are configured the enigma host shortcut in your ~.ssh/config file.
Needed a new strategy
The previous strategy works and I had been very comfortable with it. However, at times you might forget to request a node from the cluster to do your work interactively. This is specially true for me when I only plan on using a few git commands. But when many users forget this, it becomes a problem and our cluster manager had to send us a reminder:
*ALWAYS* work on a cluster node rather than on enigma2.
Enigma2 is a *single machine* with many people trying to use it to gain access to the cluster.
Even for tar commands, cp commands, wc commands ... first qrsh to a node.
Profiles in iTerm2
Just a few days after we got this reminder, I decided to take a look at the iTerm2 profile menu. There are plenty of options for customizing your terminal, but the ones I mainly ended up using are:
Working directory->Directory :-> choose a directory in my laptop. Generally the location of mygitrepository for version controlling the code of a given project.Command->Send text at start-> an alias from my local.bashrcfile as shown in the image below (the alias is qr).
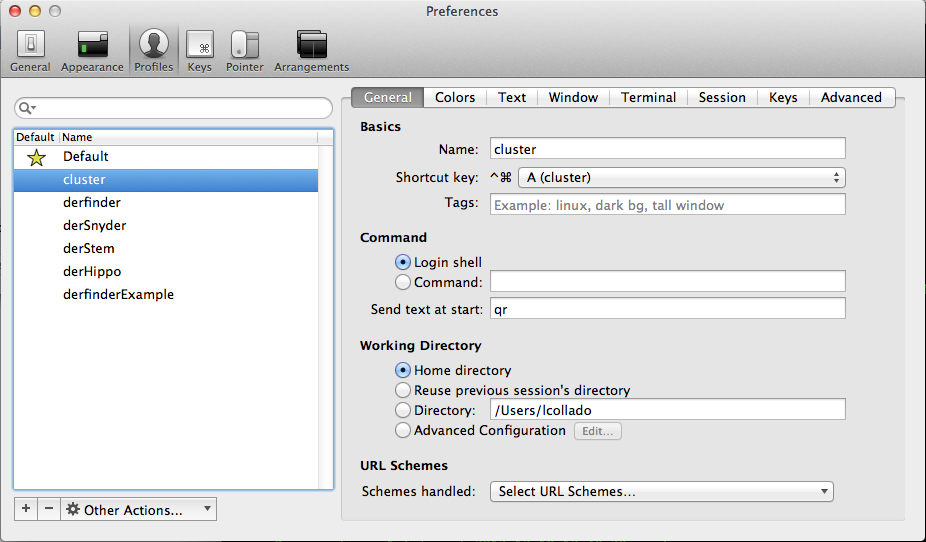
The first case above is nice, but the real power comes from the second case. Since I can pretty much evaluate any command, I asked myself if I could set up a profile that automatically logs in to the cluster? Can it also request a node interactively? And even go to my project directory?
cluster qr alias
I then remembered that Samuel Younkin explained to us how to set up qrsh to automatically change the directory to the directory from which you invoked qrsh (Younkin, 2013). I modified things a little bit and saved this qrsh version as the qr alias on the cluster:
## In cluster .bashrc file
## change dir automatically when using qrsh
## Details: https://github.com/rkostadi/BiocHopkins/wiki/Useless-Tips-&-Code-Snippets
if [ -f ~/.bash_pwd ]; then
source ~/.bash_pwd
rm ~/.bash_pwd
fi
alias qr='echo "cd $PWD" > ~/.bash_pwd; history -w; qrsh'
Note that I tried using the qrsh -ac option, but couldn’t get to pass a variable. Doing so in theory would remove the need to create the .bash_pwd temporary file.
ssh and change dir in one command
Then googling I found how to ssh and change directory in one command (Frosty, 2009):
ssh -t enigma 'cd /very/complicated/path/to/projectDir/; bash'
The problem I soon encountered was that I couldn’t qrsh right after because the command was not been found. Some setup files are not been read even after using bash -l like shown here:
ssh -t enigma 'cd /very/complicated/path/to/projectDir/; bash -l'
local qr alias
With our cluster administrator’s help, I was finally able to find how to do all of this in a single command:
## Requires the code by Sam Younkin to work (or the version I modified)
ssh -t enigma 'cd /very/complicated/path/to/projectDir/; source /etc/profile; echo "cd $PWD" > ~/.bash_pwd; history -w; qrsh'
Finally, I created the qr alias in my local machine. This alias:
- performs the ssh connection to the login machine of our cluster (enigma2),
- then changes the directory to the project directory,
- loads the necessary setup files so qrsh can be found on the path,
- creates the temporary .bash_pwd file (you could even make this more concise by echoing the directory of interest to the temp file),
- saves the command history as recommended by Samuel Younkin,
- requests an interactive node (with your default settings) by executing qrsh.
## In local .bashrc file
# qrsh
alias qr="ssh -t enigma 'cd /very/complicated/path/to/projectDir/; source /etc/profile; echo \"cd \$PWD\" > ~/.bash_pwd; qrsh'"
Note the use of the backslash to delay the execution of $PWD. I want it to be executed on the cluster, not on my local machine.
Glory!
So now using iTerm2 I can simply use the shortcut for my cluster profile which runs the local qr alias and the next thing I have is a terminal with an interactive session on the cluster and located in my project directory. Sweet! =)
Plus I also have the cluster qr alias for doing what Samuel Younkin previously described.
Can do it better?
If you have suggestions on how to improve this, let me know!
Extra aliases
The following two aliases take your local current directory basename and use it for accessing /very/complicated/path/to/projectDir/basename. This is useful if you use an organization similar to mine:
- Projects dir
- Project 1 dir
- Project 2 dir
The full paths are different in my computer and in the cluster, but once you are in /very/complicated/path/to/projectDir/ it is all the same on both locations.
The first one runs qrsh while the second one doesn’t request a node for interactive work.
## In local .bashrc file
## qrsh-basename
alias qs='LEODIR=`basename $PWD`; ssh -t enigma "cd /very/complicated/path/to/projectDir/$LEODIR/; source /etc/profile; echo \"cd \$PWD\" > ~/.bash_pwd; history -w; qrsh"'
## basename, but no qrsh
alias qq='LEODIR=`basename $PWD`; ssh -t enigma "cd /very/complicated/path/to/projectDir/$LEODIR/; source /etc/profile; echo \"cd \$PWD\" > ~/.bash_pwd; history -w; bash"'
References
Citations made with knitcitations (Boettiger, 2013).
- Frosty, (2009) How can I ssh directly to a particular directory?. How can I ssh directly to a particular directory? - Stack Overflow http://stackoverflow.com/questions/626533/how-can-i-ssh-directly-to-a-particular-directory
- BiocHopkins. GitHub http://bit.ly/19zLZeD
- Carl Boettiger, (2013) knitcitations: Citations for knitr markdown files. http://CRAN.R-project.org/package=knitcitations
Recap
If you got lost, these are the basic modifications you need to make to your local and cluster .bashrc files.
Check the history of this post here.