Getting ready to attend rOpenSci unconf18 and probably working on tidyverse-like functions for the first time
It’s Friday 7pm and it’s been a long week with ups and downs1. But I’m enthused as I write this blog post. In less than a month from now I’ll be attending rOpenSci unconf18 and it’ll be my first time at this type of event. Yay!
Building on my streak of good news, I'm delighted to have been selected to attend @rOpenSci #Unconf18 https://t.co/Xe6lojB7TS ^_^ Also, thanks to the https://t.co/o5OwUWEaBD and @LieberInstitute for their support! I'm hoping to relay as much as I can to @LIBDrstats #rstats
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) April 11, 2018
In my self introduction to everyone attending, I mentioned that I don’t use the pipe (%>%) symbol and that I use <- for assignment.
 Recently I had my pre-unconf chat with [Stefanie Butland](https://ropensci.org/about/#team) (read more about these chats [in her great blog post](https://blog.trelliscience.com/the-value-of-welcome-part-2-how-to-prepare-40-new-community-members-for-an-unconference/)). In my notes to Stefanie before our chat I had mentioned again my lack of R piping experience and we talked about it. As we talked, it became clear that a blog post on related topics would be useful. Sure, I could have asked directly to the other unconf18 participants, but maybe others from the R community in general can chime in or benefit from reading the answers.
Recently I had my pre-unconf chat with [Stefanie Butland](https://ropensci.org/about/#team) (read more about these chats [in her great blog post](https://blog.trelliscience.com/the-value-of-welcome-part-2-how-to-prepare-40-new-community-members-for-an-unconference/)). In my notes to Stefanie before our chat I had mentioned again my lack of R piping experience and we talked about it. As we talked, it became clear that a blog post on related topics would be useful. Sure, I could have asked directly to the other unconf18 participants, but maybe others from the R community in general can chime in or benefit from reading the answers.
Coding style and git
I’m attending unconf18 with an open mind and I think of myself as someone who can be quite flexible (not with my body!) and accommodating. I’m assuming that most participants at unconf18 will use = for assignment. I’m not looking to start any discussions about the different assignment operators. Simply, I am willing to use whatever the majority uses. Just like I did in my first pull request to the blogdown package (issue 263). I was trying to follow Yihui Xie’s coding style to make his life easier and have a clean (or cleaner) git history. From Yihui’s post on this pull request I can see that he liked it.
Keeping this in mind, I think that following the coding style of others will be something I’ll do at unconf18. I haven’t really worked in any R packages with many developers actively working on the package. But I imagine that setting a common coding style will minimize git conflicts, and no one wants those2. I don’t know if we’ll all follow some common guidelines at unconf18. I actually imagine that it’ll be project-specific. Why? Well because you can create an R project in RStudio3 and set some defaults for the project such as:
- the number of spaces for tab
- line ending conversions
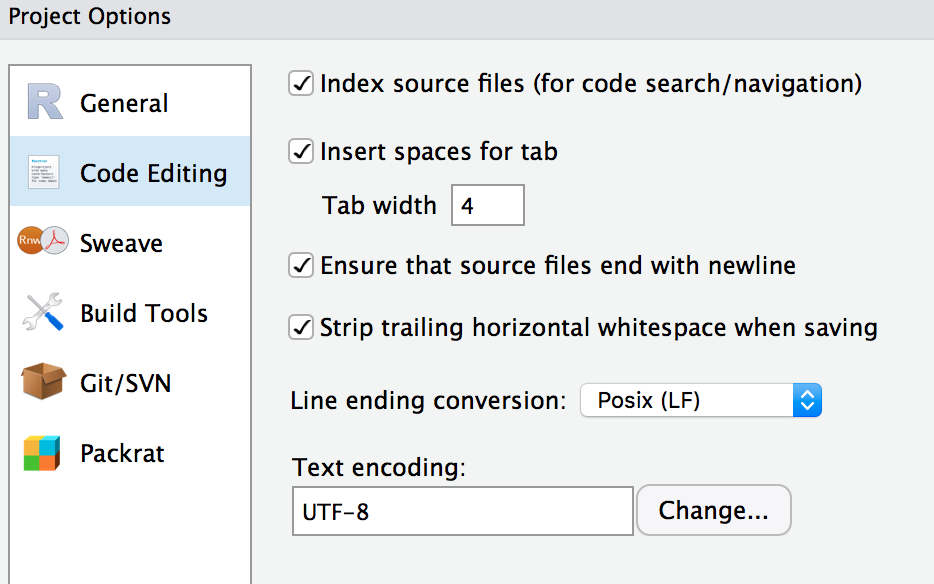
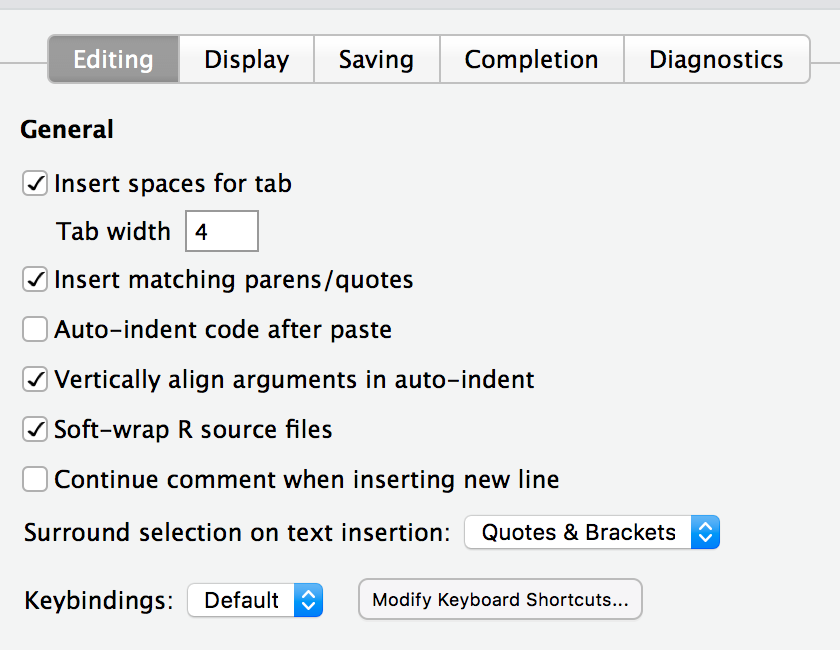
We can also set some global RStudio preferences like whether to auto-indent code after paste, length of lines.
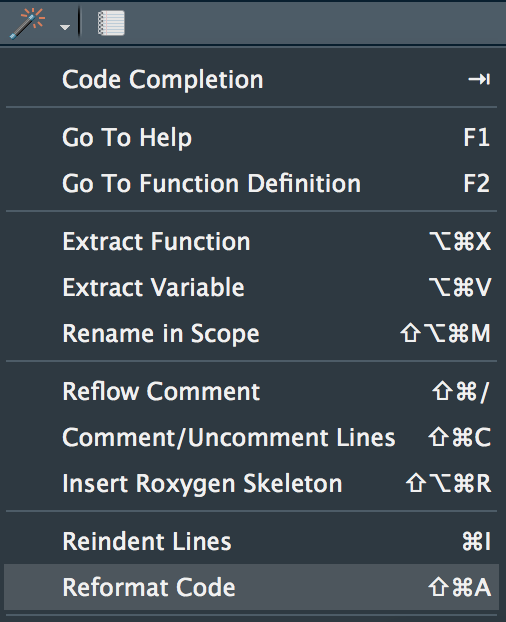
 Additionally, we can decide whether we’ll use the RStudio “wand” to *reformat code*.
Additionally, we can decide whether we’ll use the RStudio “wand” to *reformat code*.
Maybe all of this is unnecessary, maybe everyone will work in different non-overlapping functions and thus avoid git conflicts. For example, at my job sometimes I write code with = users, but we don’t work on the same lines of the code file. Later on it becomes easy to identify who wrote which line without having to use git blame (awful name, right?).
Tidyverse-like functions
So far, I think that these coding style issues are minor and will be easily dealt with. I think that we’ll all be able to adapt and instead focus on other problems (like whatever the package is trying to solve) and enjoy the experience (network, build trust as Stefanie put it).
My second concern has to do with something I imagine could require more effort: my homework before the unconf. That is, writing tidyverse-like functions. Like I’ve said, I haven’t used the R pipe %>% symbol. I’ve executed some code with it before, seeing it in many blog posts, but I’ve never actually written functions designed to be compatible with this type of logic.
If I help write a function that is not pipe-friendly, then it might not integrate nicely with other functions written by the rest of the team. It would lead to workarounds in the vignette or maybe having someone else re-factor my first function to make it pipe-friendly. Sure, I would learn from observing others make changes to my code. But I want to take advantage as much as I can from my experience at rOpenSci unconf4!
Since I don’t really use %>%, I’m unfamiliar with many things related to pipe-friendly (tidyverse-like) functions. For example:
- Do you document them differently? Like make a single Rd file for multiple functions. Or do you make an Rd file per function even if the example usage doesn’t involve
%>%? - I know that the first argument is important in pipe-friendly functions. But I ignore if the second and other arguments play a role or not.
- Do people use the ellipsis (
...) argument in pipe-friendly functions? With my derfinder package I ended up a very deep rabbit hole using.... I explained some of the logic in mydotsblog post (there are fair criticisms to going deep with...in the comments). - How do you write unit tests for pipe-friendly functions? Similar to how you write documentation for them, do the unit tests just test one function a time or do they test several at a time (that is the output after using
%>%)?
These and other questions could involve time getting familiar with. Time that I could spend now, before unconf18, learning and at least getting a better sense of what to expect. Maybe I’m complicating myself and worrying too much about this. I imagine that the solution will involve a combination of:
- Checking some popular tidyverse packages that use
%>%. Like the vignettes/README files and examples. - Reading more about this in a book(s): I don’t know which one though.
- Playing around a bit as a user with some of these functions. See what error messages pop up: actually I don’t know how users debug a series of functions tied together via
%>%.
Wrapping up
I’m not saying everyone should learn about these topics before unconf18. I think that we are all (well, maybe excluding some) worried about not knowing \(X\) or \(Y\) R/git/travis/testthat/usethis/etc topic before unconf18. And that will part of why it’ll be great to meet everyone in what is known to be an extremely welcoming R conference ^^ (seriously, check all the unconf17 posts!).
## And I'm done proofreading the post. Yay!
Sys.time()
## [1] "2023-07-11 16:54:21 EDT"
Acknowledgments
This blog post was made possible thanks to:
- BiocStyle (Oleś, 2023)
- blogdown (Xie, Hill, and Thomas, 2017)
- devtools (Wickham, Hester, Chang, and Bryan, 2022)
- knitcitations (Boettiger, 2021)
References
\[1\] C. Boettiger. knitcitations: Citations for ‘Knitr’ Markdown Files. R package version 1.0.12. 2021. URL: https://CRAN.R-project.org/package=knitcitations.
\[2\] A. Oleś. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.28.0. 2023. DOI: 10.18129/B9.bioc.BiocStyle. URL: https://bioconductor.org/packages/BiocStyle.
\[3\] H. Wickham, J. Hester, W. Chang, and J. Bryan. devtools: Tools to Make Developing R Packages Easier. R package version 2.4.5. 2022. URL: https://CRAN.R-project.org/package=devtools.
\[4\] Y. Xie, A. P. Hill, and A. Thomas. blogdown: Creating Websites with R Markdown. Boca Raton, Florida: Chapman and Hall/CRC, 2017. ISBN: 978-0815363729. URL: https://bookdown.org/yihui/blogdown/.
Reproducibility
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.3.1 (2023-06-16)
## os macOS Ventura 13.4
## system aarch64, darwin20
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/New_York
## date 2023-07-11
## pandoc 3.1.5 @ /opt/homebrew/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
## bibtex 0.5.1 2023-01-26 [1] CRAN (R 4.3.0)
## BiocManager 1.30.21 2023-06-10 [1] CRAN (R 4.3.0)
## BiocStyle * 2.28.0 2023-04-25 [1] Bioconductor
## blogdown 1.18 2023-06-19 [1] CRAN (R 4.3.0)
## bookdown 0.34 2023-05-09 [1] CRAN (R 4.3.0)
## bslib 0.5.0 2023-06-09 [1] CRAN (R 4.3.0)
## cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
## callr 3.7.3 2022-11-02 [1] CRAN (R 4.3.0)
## cli 3.6.1 2023-03-23 [1] CRAN (R 4.3.0)
## colorout 1.2-2 2023-05-06 [1] Github (jalvesaq/colorout@79931fd)
## crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
## devtools * 2.4.5 2022-10-11 [1] CRAN (R 4.3.0)
## digest 0.6.31 2022-12-11 [1] CRAN (R 4.3.0)
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
## evaluate 0.21 2023-05-05 [1] CRAN (R 4.3.0)
## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
## fs 1.6.2 2023-04-25 [1] CRAN (R 4.3.0)
## generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
## glue 1.6.2 2022-02-24 [1] CRAN (R 4.3.0)
## htmltools 0.5.5 2023-03-23 [1] CRAN (R 4.3.0)
## htmlwidgets 1.6.2 2023-03-17 [1] CRAN (R 4.3.0)
## httpuv 1.6.11 2023-05-11 [1] CRAN (R 4.3.0)
## httr 1.4.6 2023-05-08 [1] CRAN (R 4.3.0)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
## jsonlite 1.8.7 2023-06-29 [1] CRAN (R 4.3.0)
## knitcitations * 1.0.12 2021-01-10 [1] CRAN (R 4.3.0)
## knitr 1.43 2023-05-25 [1] CRAN (R 4.3.0)
## later 1.3.1 2023-05-02 [1] CRAN (R 4.3.0)
## lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.3.0)
## lubridate 1.9.2 2023-02-10 [1] CRAN (R 4.3.0)
## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
## mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
## miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
## pkgbuild 1.4.2 2023-06-26 [1] CRAN (R 4.3.0)
## pkgload 1.3.2 2022-11-16 [1] CRAN (R 4.3.0)
## plyr 1.8.8 2022-11-11 [1] CRAN (R 4.3.0)
## prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.3.0)
## processx 3.8.2 2023-06-30 [1] CRAN (R 4.3.0)
## profvis 0.3.8 2023-05-02 [1] CRAN (R 4.3.0)
## promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.3.0)
## ps 1.7.5 2023-04-18 [1] CRAN (R 4.3.0)
## purrr 1.0.1 2023-01-10 [1] CRAN (R 4.3.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
## Rcpp 1.0.10 2023-01-22 [1] CRAN (R 4.3.0)
## RefManageR 1.4.0 2022-09-30 [1] CRAN (R 4.3.0)
## remotes 2.4.2 2021-11-30 [1] CRAN (R 4.3.0)
## rlang 1.1.1 2023-04-28 [1] CRAN (R 4.3.0)
## rmarkdown 2.23 2023-07-01 [1] CRAN (R 4.3.0)
## rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.3.0)
## sass 0.4.6.9000 2023-05-06 [1] Github (rstudio/sass@f248fe5)
## sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
## shiny 1.7.4 2022-12-15 [1] CRAN (R 4.3.0)
## stringi 1.7.12 2023-01-11 [1] CRAN (R 4.3.0)
## stringr 1.5.0 2022-12-02 [1] CRAN (R 4.3.0)
## timechange 0.2.0 2023-01-11 [1] CRAN (R 4.3.0)
## urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.3.0)
## usethis * 2.2.1 2023-06-23 [1] CRAN (R 4.3.0)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.3.0)
## xfun 0.39 2023-04-20 [1] CRAN (R 4.3.0)
## xml2 1.3.4 2023-04-27 [1] CRAN (R 4.3.0)
## xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
## yaml 2.3.7 2023-01-23 [1] CRAN (R 4.3.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
-
Feeling welcomed can be hard… oh well. ↩︎
-
We could talk about git for a long time too. But I hope that I’ll get by with some git push, git pull, and maybe git branch. ↩︎
-
It’s one of the sponsors http://unconf18.ropensci.org/#sponsors and well, probably want most will be using since it has such nice tools for writing R packages. ↩︎
-
Specially since most only attend one unconf, I think. So it’s different from other conferences that you can experience multiple years and network with the group across a longer period of time: that’s what I’ve done with the Bioconductor meetings. ↩︎