How to reduce the size of a large GitHub repo
This work was done by Leo with Erik Nelson and Ryan Miller. Thank you for your time and input!
GitHub repository is huge! 😱 What do I do now?
Our story
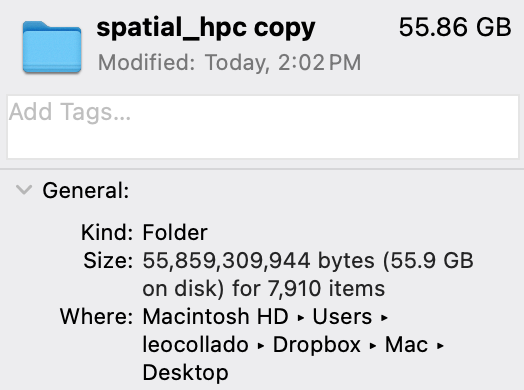
That’s how things start. In our case, we had a repository that would download over 22.6 GB of data and use 55.9 GB of disk space. Note: This repository is currently (April 15 2024) private, but will be made public soon.
% git clone git@github.com:LieberInstitute/spatial_hpc.git
Cloning into 'spatial_hpc'...
remote: Enumerating objects: 18189, done.
remote: Counting objects: 100% (1999/1999), done.
remote: Compressing objects: 100% (1026/1026), done.
remote: Total 18189 (delta 963), reused 1981 (delta 950), pack-reused 16190
Receiving objects: 100% (18189/18189), 22.64 GiB | 12.90 MiB/s, done.
Resolving deltas: 100% (9734/9734), done.
Updating files: 100% (6667/6667), done.
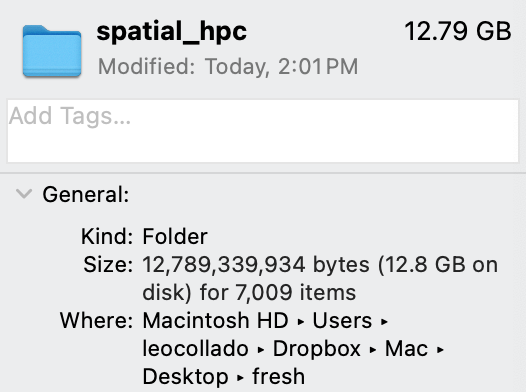
We were able to reduce it such that it now downloads only 5.1 GB of data and takes 12.8 GB of disk space. While this could still be considered large by others, it’s much easier to download and tries to balance publicly providing some plots versus none.
% git clone git@github.com:LieberInstitute/spatial_hpc.git
Cloning into 'spatial_hpc'...
remote: Enumerating objects: 18451, done.
remote: Counting objects: 100% (8112/8112), done.
remote: Compressing objects: 100% (2851/2851), done.
remote: Total 18451 (delta 4129), reused 7963 (delta 4006), pack-reused 10339
Receiving objects: 100% (18451/18451), 5.13 GiB | 11.41 MiB/s, done.
Resolving deltas: 100% (9433/9433), done.
Updating files: 100% (5784/5784), done.
BFG Repo-Cleaner and removed all files over 10 MB except for two files that we wanted to keep.
Steps by steps
Get your workspace ready
We mostly work on a high performance computing (HPC) environment, aka a compute cluster, called JHPCE. In general for our projects, we have very large files that we do not version control. That is, we ignore them thanks to one or multiple .gitignore files. Given the complexity of differentiating version controlled files from ignored files, my recommendation is to work on your work computer (in my case, a laptop).
With this in mind, we did a fresh clone of the repository as shown previously.
% git clone git@github.com:LieberInstitute/spatial_hpc.git
Cloning into 'spatial_hpc'...
remote: Enumerating objects: 18189, done.
remote: Counting objects: 100% (1999/1999), done.
remote: Compressing objects: 100% (1026/1026), done.
remote: Total 18189 (delta 963), reused 1981 (delta 950), pack-reused 16190
Receiving objects: 100% (18189/18189), 22.64 GiB | 12.90 MiB/s, done.
Resolving deltas: 100% (9734/9734), done.
Updating files: 100% (6667/6667), done.
This is also the first step that BFG recommends that you do. In general, you want to make sure that you have a pristine copy that you can use as a fallback option in case anything goes wrong, and a working copy.
--mirror option, although as you’ll see later, we did need to have a full copy of our files in our computer. So I actually recommend that you do not use --mirror.
We also downloaded BFG’s latest version: currently that is v1.14.0. This downloads the bfg-1.14.0.jar file. On a macOS computer, you also need to install Java from https://www.java.com/en/download/. As my macOS computer is from the M series system (ARM64), I did have to download the right version for it instead of the one from the big green button that downloads Java for macOS Intel (incompatible with my computer!).

You can then test that it’s all working as intended with this command:
% java -jar ~/Desktop/bfg-1.14.0.jar
bfg 1.14.0
Usage: bfg [options] [<repo>]
-b, --strip-blobs-bigger-than <size>
strip blobs bigger than X (eg '128K', '1M', etc)
-B, --strip-biggest-blobs NUM
strip the top NUM biggest blobs
-bi, --strip-blobs-with-ids <blob-ids-file>
strip blobs with the specified Git object ids
-D, --delete-files <glob>
delete files with the specified names (eg '*.class', '*.{txt,log}' - matches on file name, not path within repo)
--delete-folders <glob> delete folders with the specified names (eg '.svn', '*-tmp' - matches on folder name, not path within repo)
--convert-to-git-lfs <value>
extract files with the specified names (eg '*.zip' or '*.mp4') into Git LFS
-rt, --replace-text <expressions-file>
filter content of files, replacing matched text. Match expressions should be listed in the file, one expression per line - by default, each expression is treated as a literal, but 'regex:' & 'glob:' prefixes are supported, with '==>' to specify a replacement string other than the default of '***REMOVED***'.
-fi, --filter-content-including <glob>
do file-content filtering on files that match the specified expression (eg '*.{txt,properties}')
-fe, --filter-content-excluding <glob>
don't do file-content filtering on files that match the specified expression (eg '*.{xml,pdf}')
-fs, --filter-content-size-threshold <size>
only do file-content filtering on files smaller than <size> (default is 1048576 bytes)
-p, --protect-blobs-from <refs>
protect blobs that appear in the most recent versions of the specified refs (default is 'HEAD')
--no-blob-protection allow the BFG to modify even your *latest* commit. Not recommended: you should have already ensured your latest commit is clean.
--private treat this repo-rewrite as removing private data (for example: omit old commit ids from commit messages)
--massive-non-file-objects-sized-up-to <size>
increase memory usage to handle over-size Commits, Tags, and Trees that are up to X in size (eg '10M')
<repo> file path for Git repository to clean
Diagnose
Once you have BFG installed as well as a working clone (copy) of your GitHub repository, you can proceed to identify large files. Say files larger than 100 MB. That is the strategy that you typically use when you version controlled large files and made other smaller commits, and now cannot git push (upload files) to GitHub. In our case, we had no files over 100 MB.
% cd ~/Desktop/spatial_hpc
% java -jar ~/Desktop/bfg-1.14.0.jar --strip-blobs-bigger-than '100M' .git
Using repo : /Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git
Scanning packfile for large blobs: 18215
Scanning packfile for large blobs completed in 89 ms.
Warning : no large blobs matching criteria found in packfiles - does the repo need to be packed?
So we then proceeded to check files over 10 MB (we also checked files over 50 MB but we won’t talk about that analysis in this blog post). BFG provides a lot of output as shown below. BFG told us that we had 885 “dirty files”. These are files over 10 MB (the threshold value we specified) that are present in our latest version on GitHub. It also found “clean files” over 10 MB, which are files that at some point we had version controlled but then deleted them. So they only exist in the git history in case we need to go back in time, but are not visible from the landing page on GitHub (though you can use GitHub to navigate the git history if you want to).
% java -jar ~/Desktop/bfg-1.14.0.jar --strip-blobs-bigger-than '10M' .git
Using repo : /Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git
Scanning packfile for large blobs: 18215
Scanning packfile for large blobs completed in 91 ms.
Found 1130 blob ids for large blobs - biggest=97378037 smallest=10491618
Total size (unpacked)=29298335089
Found 6108 objects to protect
Found 4 commit-pointing refs : HEAD, refs/heads/main, refs/remotes/origin/HEAD, refs/remotes/origin/main
Protected commits
-----------------
These are your protected commits, and so their contents will NOT be altered:
* commit f5f9a19e (protected by 'HEAD') - contains 885 dirty files :
- code/enrichment_analysis/project_all/score/snRNAseq_aggregated_cpm.tsv (16.4 MB)
- code/spot_deconvo/cell2location/02_registration_HE_broad_class.log (16.3 MB)
- ...
WARNING: The dirty content above may be removed from other commits, but as
the *protected* commits still use it, it will STILL exist in your repository.
Details of protected dirty content have been recorded here :
/Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git.bfg-report/2024-04-11/09-51-45/protected-dirt/
If you *really* want this content gone, make a manual commit that removes it,
and then run the BFG on a fresh copy of your repo.
Cleaning
--------
Found 1561 commits
Cleaning commits: 100% (1561/1561)
Cleaning commits completed in 1,197 ms.
Updating 2 Refs
---------------
Ref Before After
----------------------------------------------
refs/heads/main | f5f9a19e | ba48bc2c
refs/remotes/origin/main | f5f9a19e | ba48bc2c
Updating references: 100% (2/2)
...Ref update completed in 12 ms.
Commit Tree-Dirt History
------------------------
Earliest Latest
| |
..DDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD
D = dirty commits (file tree fixed)
m = modified commits (commit message or parents changed)
. = clean commits (no changes to file tree)
Before After
-------------------------------------------
First modified commit | 545cd363 | fcc2538e
Last dirty commit | ead4cbed | 600580dd
Deleted files
-------------
Filename Git id
----------------------------------------------------------------------------------------------------------------------------
02_registration_HE_broad_class.log | de11acf2 (12.9 MB), 33ae1f4d (16.3 MB)
02_registration_HE_layer.log | ba6d1c7e (19.6 MB), 279e5354 (19.6 MB)
2743.png | c734cb7d (11.3 MB)
335.png | 602fc07e (18.8 MB)
6423.png | 7071add9 (15.4 MB)
6432.png | 9b9d7fa6 (13.2 MB)
6471.png | 0f61bfd7 (17.5 MB)
6522.png | 573fab2b (19.8 MB)
8325.png | 9cf4ff3e (22.0 MB)
8492.png | 0a128ff1 (10.2 MB)
8667.png | 014ee77a (15.4 MB)
AC005837.4.pdf | 0b12e443 (32.4 MB)
AC106795.2.pdf | 60dba623 (32.4 MB), ba037e7f (32.5 MB)
AC108863.2.pdf | 56eb980a (32.4 MB)
ACAP1_DOWN.pdf | 584afb1f (32.5 MB)
...
In total, 5160 object ids were changed. Full details are logged here:
/Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git.bfg-report/2024-04-11/09-51-45
BFG run is complete! When ready, run: git reflog expire --expire=now --all && git gc --prune=now --aggressive
Among the output of BFG was a CSV file which listed the 885 “dirty files”.
% ls -lh /Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git.bfg-report/2024-04-11/09-51-45/protected-dirt/
total 132K
-rw-r--r-- 1 leocollado staff 132K Apr 11 09:51 f5f9a19e-HEAD.csv
With R, we can read that CSV file and help us diagnose our situation. We recommend that you inspect this CSV file, which will be ordered by file path.
## Read in the CSV file with R
ten <- read.csv("f5f9a19e-HEAD.csv", header = FALSE)
## How many "dirty files" do we have?
nrow(ten)
## [1] 885
## What's the format of this CSV file?
head(ten)
## V1 V2 V3
## 1 e00fd6349fcabdd740b396b230a74f9f0d136f2e DELETE executable
## 2 33ae1f4da994efaad1538c6d740ac22fb6f0aab4 DELETE regular-file
## 3 279e53549e75b881fb7b063f05211fde529c17fb DELETE regular-file
## 4 8e1c82f0c40758394abfc807a3fcc6094fef329d DELETE regular-file
## 5 f41fa6e0e3e6a25b390aa33a4d4fa3c3b493d97a DELETE regular-file
## 6 89f9bb0104d49d7584d712a8cde2e9a580356291 DELETE regular-file
## V4
## 1 code/enrichment_analysis/project_all/score/snRNAseq_aggregated_cpm.tsv
## 2 code/spot_deconvo/cell2location/02_registration_HE_broad_class.log
## 3 code/spot_deconvo/cell2location/02_registration_HE_layer.log
## 4 plots/02_build_spe/ReferenceMapping.pdf
## 5 plots/02_build_spe/ReferenceMapping_erik.pdf
## 6 plots/02_build_spe/prearrangedAllSamples.pdf
## V5 V6
## 1 17229010 NA
## 2 17115444 NA
## 3 20518064 NA
## 4 54308266 NA
## 5 54308266 NA
## 6 25909369 NA
The fourth column specifies the file path and the fifth column specifies the file size in bytes. Given that this project was organized according to our template_project, Erik (who has been heavily involved in this project) could easily diagnose whether some of the listed files were important to share or whether they were older exploration results.
## Sort them by file size if you want to
# ten <- ten[order(ten$V5, decreasing = TRUE), ]
## Check file size distribution in MB (1 MB = 1024^2 bytes)
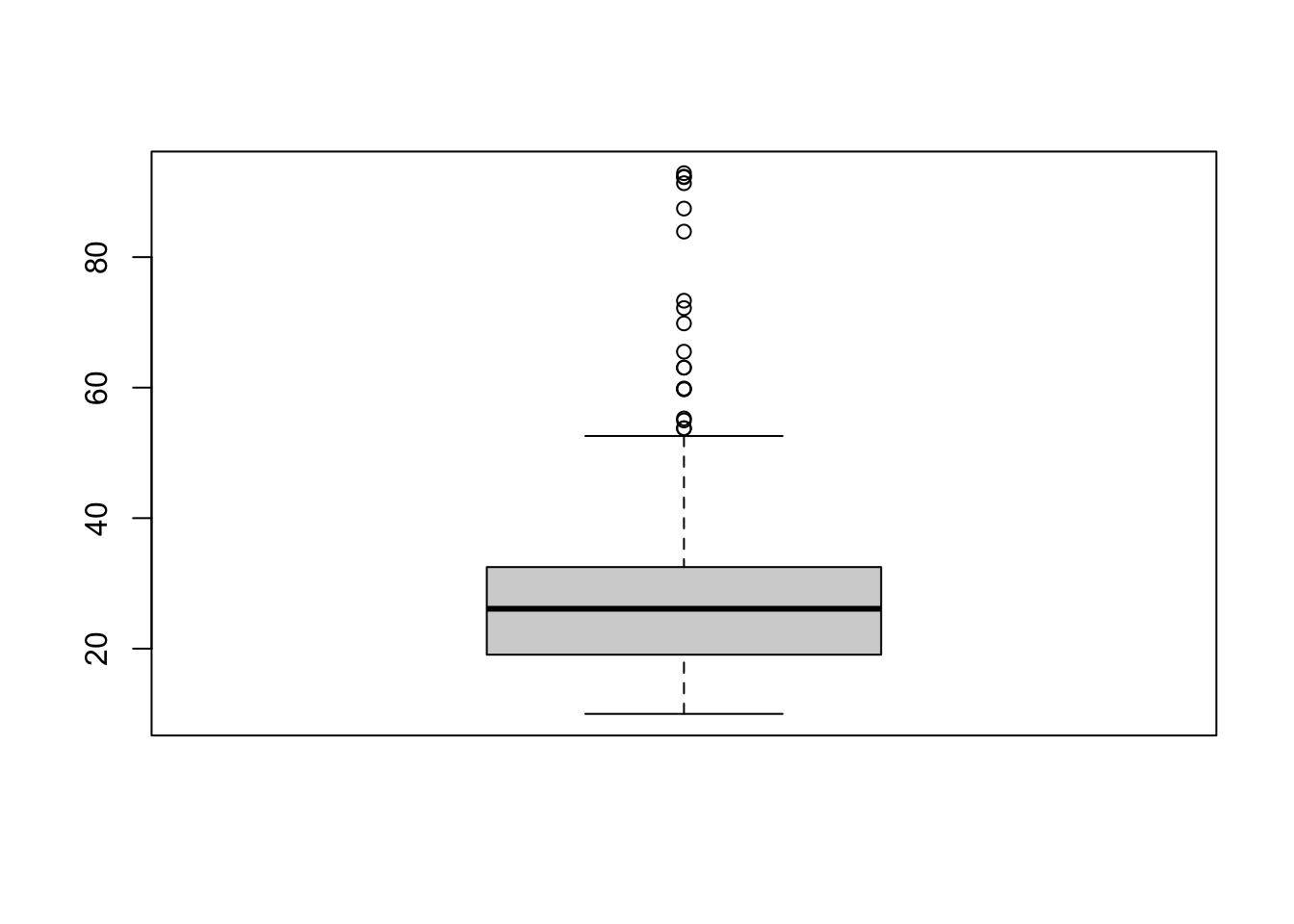
summary(ten$V5 / 1024^2)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 10.01 19.08 26.12 26.31 32.50 92.87
boxplot(ten$V5 / 1024^2)
## Explore frequency of file sizes rounded to 1 digit
barplot(table(round(ten$V5 / 1024^2, 1)))
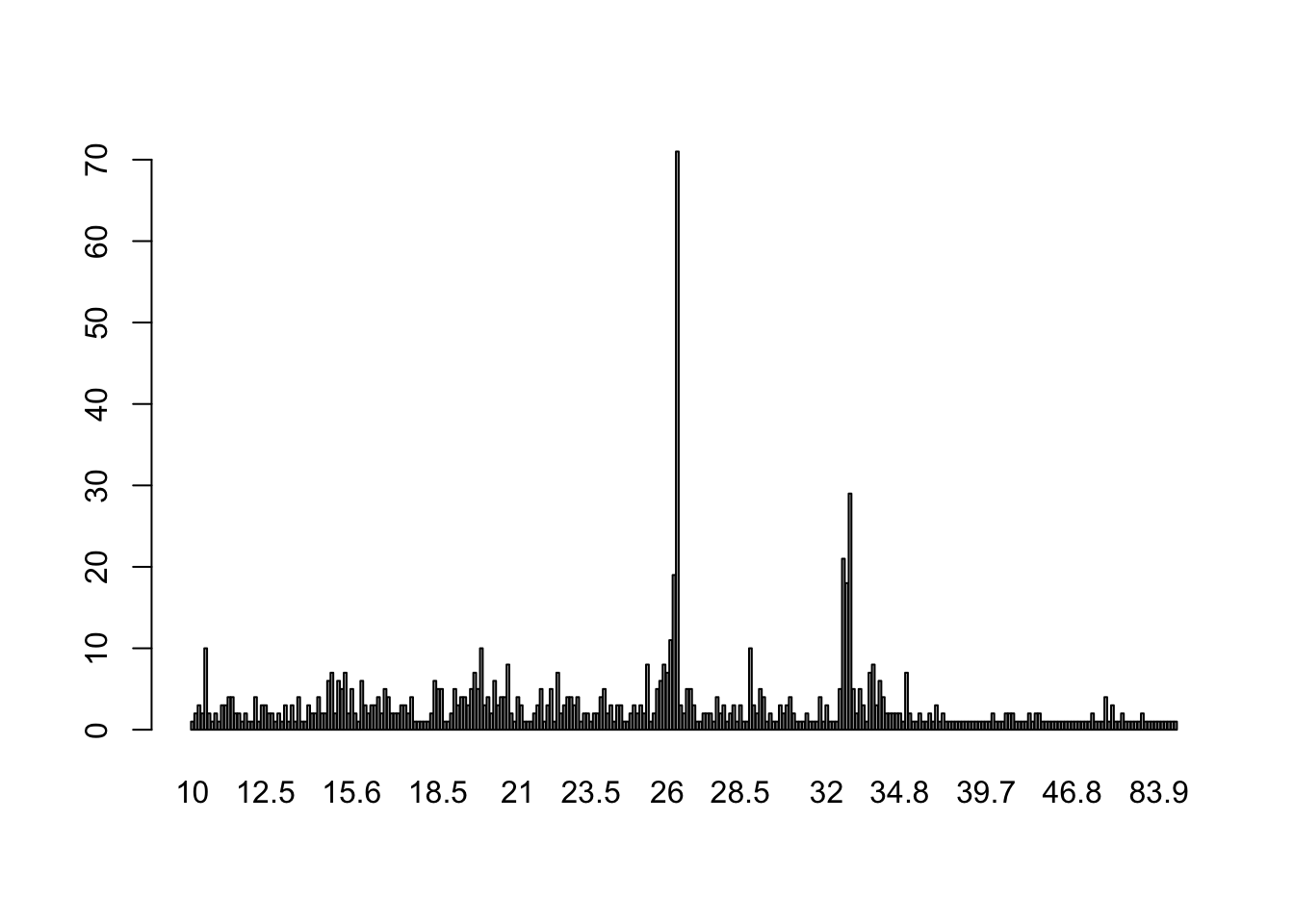
## How much total space do these "dirty files" use?
sum(ten$V5 / 1024^2) ## In MB
## [1] 23285.45
sum(ten$V5 / 1024^3) ## In GB
## [1] 22.73969
A question Erik posed was: is it mostly plots? In general, you could use file extensions if you wanted. But in our case, thanks to template_project, we can check what is the main directory for each file.
## Extract main directory from the file paths
x <- dirname(ten$V4)
x <- gsub("/.*", "", x)
## Explore how many files we have
table(x)
## x
## code plots processed-data raw-data snRNAseq_hpc
## 3 505 368 2 7
We could then explore some of the odd cases.
## Visual inspection determined that we were OK deleting these files
ten[x == "snRNAseq_hpc", ]
## V1 V2 V3
## 879 f0d8bb12bbadda84ae3e98f431083e61620869b2 DELETE executable
## 880 68b61e75d861f5d960a96cbdcad7bb5c79a3e747 DELETE regular-file
## 881 f7588d013685ebe907c972afd0aaf0935816df28 DELETE regular-file
## 882 771e792004afef8ed2cc986eb9766b01bad4db4a DELETE regular-file
## 883 204d559465dba1489955d5988a879c6cbb91a5b7 DELETE regular-file
## 884 f5bb06176473e8112d00a1d355ccd048f4dc46c1 DELETE regular-file
## 885 605957a9d3a9b2f2ed8d021dcf2c1482048f4f9a DELETE regular-file
## V4 V5 V6
## 879 snRNAseq_hpc/plots/02_qc/qc_plots.pdf 20223555 NA
## 880 snRNAseq_hpc/plots/QC_auto.pdf 41669358 NA
## 881 snRNAseq_hpc/plots/QC_semiauto.pdf 16580972 NA
## 882 snRNAseq_hpc/plots/QC_semiauto2.pdf 33358656 NA
## 883 snRNAseq_hpc/plots/build_sce/UMAP_corrected_by_sample.pdf 18167086 NA
## 884 snRNAseq_hpc/plots/build_sce/UMAPs_unccorrected.pdf 18047630 NA
## 885 snRNAseq_hpc/plots/build_sce/UMAPs_unccorrected_filtered.pdf 16767377 NA
## Same thing for these large "code" files (2 logs and one file that is not
## needed to run the actual code and thus reproduce results)
ten[x == "code", ]
## V1 V2 V3
## 1 e00fd6349fcabdd740b396b230a74f9f0d136f2e DELETE executable
## 2 33ae1f4da994efaad1538c6d740ac22fb6f0aab4 DELETE regular-file
## 3 279e53549e75b881fb7b063f05211fde529c17fb DELETE regular-file
## V4
## 1 code/enrichment_analysis/project_all/score/snRNAseq_aggregated_cpm.tsv
## 2 code/spot_deconvo/cell2location/02_registration_HE_broad_class.log
## 3 code/spot_deconvo/cell2location/02_registration_HE_layer.log
## V5 V6
## 1 17229010 NA
## 2 17115444 NA
## 3 20518064 NA
## These are important sample information files we do want to keep.
ten[x == "raw-data", ]
## V1 V2 V3
## 877 f44d0d43bd507c0e33e9267ae1ba00fda71acf66 DELETE regular-file
## 878 a3dd0e6f6056de3c11b4f3804b5ee40b39eabef4 DELETE regular-file
## V4
## 877 raw-data/FASTQ/2022-12-07_NovaSeq_33v_scp/Visium_HPC_Round1-10_092622_Master_SCP.xlsx
## 878 raw-data/sample_info/Visium_HPC_Round3-8_033022_SCP.xlsx
## V5 V6
## 877 37226953 NA
## 878 34649463 NA
Remove dirty files
Once we have identified which files we no longer want to distribute through GitHub, we can proceed to eliminate them from the git history in our local working copy. Before we can use BFG, we first have to clean these “dirty files”. The first step is to remove them from the list of version controlled files using the git rm command. However, we do want to keep a copy of these files. So we use the --cached option to do so.
## Change directory to the location of your local copy
setwd("~/Desktop/spatial_hpc/")
## Create a list of commands
cmds <- paste0("git rm --cached ", ten[x != "raw-data",]$V4)
## Loop over and execute the commands with system()
for (i in seq_along(cmds)) {
message(i, ": ", Sys.time(), " ", cmds[i])
system(cmds[i])
}
Here’s how one of these commands would look like. We used system() to actually run these commands on my macOS computer.
cmds[1]
## [1] "git rm --cached code/enrichment_analysis/project_all/score/snRNAseq_aggregated_cpm.tsv"
At this point, with git rm we have told git to no longer version control these 880+ files. To avoid the mistake of version controlling them again, we then added them to the main .gitignore file using readLines() and writeLines().
## Update our main .gitignore file with the list of files
## we no longer want to version control.
gitignore <- readLines(".gitignore")
gitignore <- c(gitignore, ten[x != "raw-data",]$V4)
writeLines(gitignore, ".gitignore")
At this point, if you use git status, you’ll see the long list of files marked for removal from version control.
% git status
On branch main
Your branch is ahead of 'origin/main' by 1 commit.
(use "git push" to publish your local commits)
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
deleted: plots/02_build_spe/ReferenceMapping.pdf
deleted: plots/02_build_spe/ReferenceMapping_erik.pdf
deleted: plots/02_build_spe/prearrangedAllSamples.pdf
## and many more lines
## Note that you should not see any untracked files listed at this step.
Next, you have to make a git commit to finally unprotect these “dirty files”.
git commit -am "Unprotecting files over 10 Mb so we can remove them from git history with BFG"
Editing the git history
Now that we no longer have “dirty files” over 10 MB (except for the 2 Excel files we decided to keep), we can now run BFG again. At this point, out of precaution, I did make a full copy of my local clone (copy) of the repository. Though in retrospect, you likely want to do this before you even run BFG the very first time. That’s because BFG will detect that it has been used before on a given git repository and will prune repo before proceeding which took about 6 minutes for us.
% date
Thu Apr 11 11:27:06 EDT 2024
% java -jar ~/Desktop/bfg-1.14.0.jar --strip-blobs-bigger-than '10M' .git
Using repo : /Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git
This repo has been processed by The BFG before! Will prune repo before proceeding - to avoid unnecessary cleaning work on unused objects...
date
Completed prune of old objects - will now proceed with the main job!
Scanning packfile for large blobs: 42927
Scanning packfile for large blobs completed in 672 ms.
Found 1130 blob ids for large blobs - biggest=97378037 smallest=10491618
Total size (unpacked)=29298335089
Found 5225 objects to protect
Found 4 commit-pointing refs : HEAD, refs/heads/main, refs/remotes/origin/HEAD, refs/remotes/origin/main
Protected commits
-----------------
These are your protected commits, and so their contents will NOT be altered:
* commit 3d58fd38 (protected by 'HEAD') - contains 2 dirty files :
- raw-data/FASTQ/2022-12-07_NovaSeq_33v_scp/Visium_HPC_Round1-10_092622_Master_SCP.xlsx (35.5 MB)
- raw-data/sample_info/Visium_HPC_Round3-8_033022_SCP.xlsx (33.0 MB)
WARNING: The dirty content above may be removed from other commits, but as
the *protected* commits still use it, it will STILL exist in your repository.
Details of protected dirty content have been recorded here :
/Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git.bfg-report/2024-04-11/11-33-47/protected-dirt/
If you *really* want this content gone, make a manual commit that removes it,
and then run the BFG on a fresh copy of your repo.
Cleaning
--------
Found 1563 commits
Cleaning commits: 100% (1563/1563)
Cleaning commits completed in 1,390 ms.
Updating 1 Ref
--------------
Ref Before After
-------------------------------------
refs/heads/main | 3d58fd38 | d598aefb
Updating references: 100% (1/1)
...Ref update completed in 12 ms.
Commit Tree-Dirt History
------------------------
Earliest Latest
| |
...........................................................D
D = dirty commits (file tree fixed)
m = modified commits (commit message or parents changed)
. = clean commits (no changes to file tree)
Before After
-------------------------------------------
First modified commit | 0255622c | 35eacffa
Last dirty commit | 0255622c | 35eacffa
Deleted files
-------------
Filename Git id
----------------------------------------------------------------------------------------------------------------------------
02_registration_HE_broad_class.log | de11acf2 (12.9 MB), 33ae1f4d (16.3 MB)
02_registration_HE_layer.log | ba6d1c7e (19.6 MB), 279e5354 (19.6 MB)
2743.png | c734cb7d (11.3 MB)
335.png | 602fc07e (18.8 MB)
6423.png | 7071add9 (15.4 MB)
6432.png | 9b9d7fa6 (13.2 MB)
6471.png | 0f61bfd7 (17.5 MB)
6522.png | 573fab2b (19.8 MB)
8325.png | 9cf4ff3e (22.0 MB)
8492.png | 0a128ff1 (10.2 MB)
8667.png | 014ee77a (15.4 MB)
AC005837.4.pdf | 0b12e443 (32.4 MB)
AC106795.2.pdf | 60dba623 (32.4 MB), ba037e7f (32.5 MB)
AC108863.2.pdf | 56eb980a (32.4 MB)
ACAP1_DOWN.pdf | 584afb1f (32.5 MB)
...
In total, 5168 object ids were changed. Full details are logged here:
/Users/leocollado/Dropbox/Mac/Desktop/spatial_hpc/.git.bfg-report/2024-04-11/11-33-47
BFG run is complete! When ready, run: git reflog expire --expire=now --all && git gc --prune=now --aggressive
% date
Thu Apr 11 11:33:49 EDT 2024
As we can see on the output above, we do have 2 “dirty files”, but we knew about them. We can now proceed to run the commands BFG tells us to use to actually edit the git history.
git reflog expire --expire=now --all && git gc --prune=now --aggressive
We went to get lunch while this was happening, as my recollection was that this step can take a while to run. Though I forgot to time it this time around.
Update GitHub
We now have what we want: a much smaller git repository with only the files we want version controlled. However, it’s not on GitHub and also not in our HPC computers (JHPCE for us). So we need to proceed to update GitHub first using git push (upload command). As we are altering the git history, we need to use a git push --force command otherwise GitHub would not let us do alter the history.
% git push --force
Enumerating objects: 18451, done.
Counting objects: 100% (18451/18451), done.
Delta compression using up to 10 threads
Compressing objects: 100% (6684/6684), done.
remote: fatal: pack exceeds maximum allowed size (2.00 GiB)
error: remote unpack failed: index-pack failed 14.02 MiB/s
To github.com:LieberInstitute/spatial_hpc.git
! [remote rejected] main -> main (failed)
error: failed to push some refs to 'github.com:LieberInstitute/spatial_hpc.git'
However, as you can see, we ran into the 2 GB limit GitHub has for git push commands.
Google searching the error, we easily found this excellent documentation page. It didn’t exist (or I didn’t find it) a few years ago and it was quite straightforward to understand in contrast to previous Stack Overflow answers I remember struggling with back then.
Looking at our repository on GitHub, we saw that we have a bit over 1,500 commits. So searching every 1,000 commits most likely wasn’t going to work for us as the GitHub documentation suggested given that we would most likely run again into the 2 GB upload limit. We decided to roll with 200 commits at a time hoping that this would give us a small enough set of commit IDs to make incremental git pushes, but without exceeding the 2 GB upload limit GitHub enforces.
% git log --oneline --reverse refs/heads/main | awk 'NR % 200 == 0'
d983b009 marker gene plots
df46ec6e log file for BayesSpace rerun k15, long array
066777d0 Added a README.md file and softlinks for round9 (samples 33v-36v)[33v sample (10ul) library MiSeq and NovaSeq,samples 34v-26v NovaSeq]
51ec749f rerun VNS for sample 36 with more memory
fd032a96 tangram logs 4,5,6
e9e6526f run updated code on sample 24
9572195c updated samui image stitching script to remove banding from center of the image and average pixel intensity of the image overlaps
Thanks to the commit messages, Erik and Ryan could easily detect that the commit IDs were ordered by time such that the last commit was one of the most recent ones. We thus proceeded to run each of the git push commands. Note that we used git push --force on our first commit. After this, our repository on GitHub listed only 200 commits: what we wanted to do.
% git push --force origin d983b009:refs/heads/main
Enumerating objects: 2253, done.
Counting objects: 100% (2253/2253), done.
Delta compression using up to 10 threads
Compressing objects: 100% (1009/1009), done.
Writing objects: 100% (2253/2253), 633.78 MiB | 36.74 MiB/s, done.
Total 2253 (delta 1176), reused 1987 (delta 1085), pack-reused 0 (from 0)
remote: Resolving deltas: 100% (1176/1176), done.
To github.com:LieberInstitute/spatial_hpc.git
+ f5f9a19e...d983b009 d983b009 -> main (forced update)
Some of them got close to the 2 GB limit (like 1.5 GB), but they were all fine.
## Commands shown with no output
% git push origin df46ec6e:refs/heads/main
% git push origin 066777d0:refs/heads/main
% git push origin 51ec749f:refs/heads/main
% git push origin fd032a96:refs/heads/main
% git push origin e9e6526f:refs/heads/main
% git push origin 9572195c:refs/heads/main
% git push
Note the last git push we did at the end, which was needed to upload the last left over commits. Also note that this step was fairly simple for us as we only have 1 development branch called main for this project, instead of working across multiple git branches. If you are working with multiple branches, you might have to do quite a bit of extra work.
Updating HPC files
Are we done yet? Close! We have what we want in our local git clone (copy) on my laptop as well as at GitHub, but not in our HPC computers (JHPCE for us). We followed this Stack Overflow solution to update files at JHPCE:
## Log in to your HPC system (JHPCE for us)
## Navigate to the directory where the data lives at JHPCE
cd /dcs04/lieber/lcolladotor/spatialHPC_LIBD4035/spatial_hpc
## Download all files from GitHub to our HPC (JHPCE)
git fetch
## Force update the "main" branch
git reset origin/main --hard
At this point, we did run into a warning because I didn’t have the permissions to delete one file.
warning: unable to unlink 'code/enrichment_analysis/project_all/score/snRNAseq_aggregated_cpm.tsv': Permission denied
As you might already be wondering, git reset origin/main --hard did erase all 880+ files (except the one I didn’t have permissions for) over 10 Mb that we deleted from git and GitHub. But we wanted to have them at JHPCE. This is where having a local copy in our laptop becomes handy. We then used rsync to upload back these files to our HPC computers from my laptop.
## From your computer (my laptop in this case), upload the local files you have
## that were deleted from git / GitHub that you want to have in your HPC system
rsync -avhP . lcollado@jhpce-transfer01.jhsph.edu:/dcs04/lieber/lcolladotor/spatialHPC_LIBD4035/spatial_hpc
However, this process broke some file permissions. As we already have a script for updating permissions, I simply re-ran it at JHPCE with:
## Log in to your HPC system (JHPCE for us)
## Navigate to the directory where the data lives at JHPCE
$ cd /dcs04/lieber/lcolladotor/spatialHPC_LIBD4035/spatial_hpc
## Run script for updating permissions
$ sh /dcs04/lieber/lcolladotor/_jhpce_org_LIBD001/update_permissions_spatialteam.sh .
**** Updating permissions for . ****
Thu Apr 11 01:49:01 PM EDT 2024
**** Note that warning/error messages are expected for files and directories that you are not the owner of.
The expected warning/error messages are:
'chgrp: changing group of ‘some_JHPCE_file_path’: Operation not permitted'
or 'chmod: changing permissions of ‘some_JHPCE_file_path’: Operation not permitted'.
If for many files you are not the owner (creator of), you will get lots of these warning/error messages, this is expected!
Error or warnings with another syntax are likely real. ****
You will need to re-run this script anytime you upload files to JHPCE through Cyberduck / WinSCP as they break the ACLs.
Every new team member on a given project will likely also need to run this script once.
For more details about setting permissions at JHPCE using ACLs, please check https://lcolladotor.github.io/bioc_team_ds/organizing-your-work.html#setting-jhpce-file-permissions.
This message will be displayed for 90 seconds before the script proceeds.
That way you will have time enough to read it and/or copy it.
**** Setting read (R), write (W), and execute (X) permissions for hickslab ****
Thu Apr 11 01:50:36 PM EDT 2024
## truncated output
**** Setting the group sticky bit ****
Thu Apr 11 03:05:53 PM EDT 2024
**** Checking the nfs4 (ACLs) settings ****
Thu Apr 11 03:07:11 PM EDT 2024
# file: .
A::OWNER@:rwaDxtTcCy
A::GROUP@:rxtcy
A:g:lieber_lcolladotor@cm.cluster:rwaDxtcy
A:g:lieber_marmaypag@cm.cluster:rwaDxtcy
A:g:hickslab@cm.cluster:rxtcy
A::EVERYONE@:rxtcy
A:fdi:OWNER@:rwaDxtTcCy
A:fdi:GROUP@:rwaDxtcy
A:fdig:lieber_lcolladotor@cm.cluster:rwaDxtcy
A:fdig:lieber_marmaypag@cm.cluster:rwaDxtcy
A:fdig:hickslab@cm.cluster:rwaDxtcy
A:fdi:EVERYONE@:tcy
Now everyone else involved in this team project can keep working with the files at JHPCE and GitHub.
Finishing step
Erik will check if any of the 880+ files over 10 MB that we deleted from the git history (and currently have at JHPCE but are not on GitHub ) are actually files we should have on GitHub. If so, he’ll edit the main .gitignore accordingly (remove them from the main .gitignore ) + then git add them again.
Summary
This whole process took about three and a half hours. I hadn’t done something like this in a while so my memory wasn’t fresh. That’s why I’m writing this blog post, so we can have a good guide for the next time we have to do this.
Here are some main notes:
- Announce about a week in advance that no one should be working on the files at your HPC system (JHPCE for us): “closed for maintenance ⚠️”
- This will give people enough time to plan ahead.
- Ask people to run the update permissions script in advance.
- We got lucky this time with encountering only 1 restricted file!
- Work on a fresh
git clonein your laptop - Feel free to make a copy on your laptop before you use
BFG - Work with people who are very familiar with the contents of the repository.
- They will make crucial decisions on which files to keep or not!!
- Allocate plenty of time.
- We spent about 4 hours, but you might want to plan for 4-6 hours.
- Don’t copy paste more than 1 command into the terminal at a time.
- You might encounter errors not previously documented!
Enjoy! Best of luck with navigating similar situations.
Acknowledgments
This blog post was made possible thanks to:
- BiocStyle (Oleś, 2024)
- blogdown (Xie, Hill, and Thomas, 2017)
- knitcitations (Boettiger, 2021)
- sessioninfo (Wickham, Chang, Flight, Müller et al., 2021)
References
[1] C. Boettiger. knitcitations: Citations for 'Knitr' Markdown Files. R package version 1.0.12. 2021. URL: https://CRAN.R-project.org/package=knitcitations.
[2] A. Oleś. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.32.0. 2024. URL: https://github.com/Bioconductor/BiocStyle.
[3] H. Wickham, W. Chang, R. Flight, K. Müller, et al. sessioninfo: R Session Information. R package version 1.2.2. 2021. URL: https://CRAN.R-project.org/package=sessioninfo.
[4] Y. Xie, A. P. Hill, and A. Thomas. blogdown: Creating Websites with R Markdown. Boca Raton, Florida: Chapman and Hall/CRC, 2017. ISBN: 978-0815363729. URL: https://bookdown.org/yihui/blogdown/.
Reproducibility
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.4.0 (2024-04-24)
## os macOS Sonoma 14.5
## system aarch64, darwin20
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/New_York
## date 2024-05-23
## pandoc 3.2 @ /opt/homebrew/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## backports 1.4.1 2021-12-13 [1] CRAN (R 4.4.0)
## bibtex 0.5.1 2023-01-26 [1] CRAN (R 4.4.0)
## BiocManager 1.30.23 2024-05-04 [1] CRAN (R 4.4.0)
## BiocStyle * 2.32.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.0)
## blogdown 1.19 2024-02-01 [1] CRAN (R 4.4.0)
## bookdown 0.39 2024-04-15 [1] CRAN (R 4.4.0)
## bslib 0.7.0 2024-03-29 [1] CRAN (R 4.4.0)
## cachem 1.1.0 2024-05-16 [1] CRAN (R 4.4.0)
## cli 3.6.2 2023-12-11 [1] CRAN (R 4.4.0)
## colorout * 1.3-0.2 2024-05-03 [1] Github (jalvesaq/colorout@c6113a2)
## digest 0.6.35 2024-03-11 [1] CRAN (R 4.4.0)
## evaluate 0.23 2023-11-01 [1] CRAN (R 4.4.0)
## fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.4.0)
## generics 0.1.3 2022-07-05 [1] CRAN (R 4.4.0)
## glue 1.7.0 2024-01-09 [1] CRAN (R 4.4.0)
## highr 0.10 2022-12-22 [1] CRAN (R 4.4.0)
## htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.4.0)
## httr 1.4.7 2023-08-15 [1] CRAN (R 4.4.0)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.4.0)
## jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.4.0)
## knitcitations * 1.0.12 2021-01-10 [1] CRAN (R 4.4.0)
## knitr 1.46 2024-04-06 [1] CRAN (R 4.4.0)
## lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.4.0)
## lubridate 1.9.3 2023-09-27 [1] CRAN (R 4.4.0)
## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.4.0)
## plyr 1.8.9 2023-10-02 [1] CRAN (R 4.4.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.4.0)
## Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.4.0)
## RefManageR 1.4.0 2022-09-30 [1] CRAN (R 4.4.0)
## rlang 1.1.3 2024-01-10 [1] CRAN (R 4.4.0)
## rmarkdown 2.27 2024-05-17 [1] CRAN (R 4.4.0)
## rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.4.0)
## sass 0.4.9.9000 2024-05-03 [1] Github (rstudio/sass@9228fcf)
## sessioninfo * 1.2.2 2021-12-06 [1] CRAN (R 4.4.0)
## stringi 1.8.4 2024-05-06 [1] CRAN (R 4.4.0)
## stringr 1.5.1 2023-11-14 [1] CRAN (R 4.4.0)
## timechange 0.3.0 2024-01-18 [1] CRAN (R 4.4.0)
## xfun 0.44 2024-05-15 [1] CRAN (R 4.4.0)
## xml2 1.3.6 2023-12-04 [1] CRAN (R 4.4.0)
## yaml 2.3.8 2023-12-11 [1] CRAN (R 4.4.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────