Megadepth: efficient coverage quantification for BigWigs and BAMs
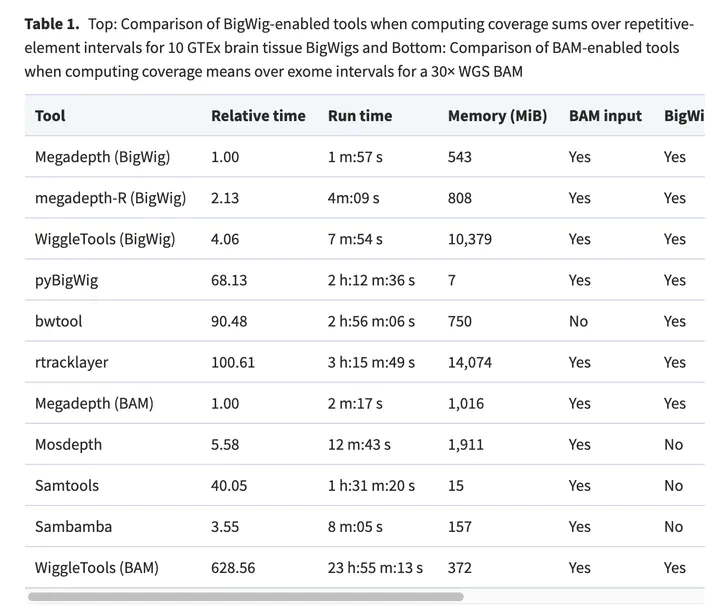 Image credit: Oxford Bioinformatics
Image credit: Oxford Bioinformatics
Abstract
Motivation. A common way to summarize sequencing datasets is to quantify data lying within genes or other genomic intervals. This can be slow and can require different tools for different input file types. Results. Megadepth is a fast tool for quantifying alignments and coverage for BigWig and BAM/CRAM input files, using substantially less memory than the next-fastest competitor. Megadepth can summarize coverage within all disjoint intervals of the Gencode V35 gene annotation for more than 19 000 GTExV8 BigWig files in approximately 1 h using 32 threads. Megadepth is available both as a command-line tool and as an R/Bioconductor package providing much faster quantification compared to the rtracklayer package. Availability and implementation: https://github.com/ChristopherWilks/megadepth, https://bioconductor.org/packages/megadepth.
Megadepth: fast quantification of interval sets w/r/t BigWigs & BAMs. Command-line & Bioconductor interfaces: https://t.co/6rq6cFhLtu. Github: https://t.co/vAcWCTrN4P. Great work, @chrisnwilks, @oyfahmed, @lcolladotor, @dyzhang32, @dnb_hopkins. 1/2 https://t.co/ogcOFYpVN2
— Ben Langmead (@BenLangmead) December 18, 2020