Splicing accuracy varies across human introns, tissues, age and disease
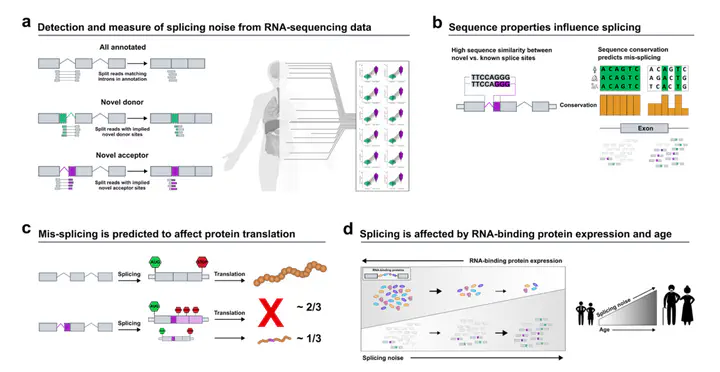 Image credit: bioRxiv
Image credit: bioRxiv
Abstract
Alternative splicing impacts most multi-exonic human genes. Inaccuracies during this process may have an important role in ageing and disease. Here, we investigate splicing accuracy using RNA-sequencing data from {\textgreater}14k control samples and 40 human body sites, focusing on split reads partially mapping to known transcripts in annotation. We show that splicing inaccuracies occur at different rates across introns and tissues and are affected by the abundance of core components of the spliceosome assembly and its regulators. We find that age is positively correlated with a global decline in splicing fidelity, mostly affecting genes implicated in neurodegenerative diseases. We find support for the latter by observing a genome-wide increase in splicing inaccuracies in samples affected with Alzheimer’s disease as compared to neurologically normal individuals. In this work, we provide an in-depth characterisation of splicing accuracy, with implications for our understanding of the role of inaccuracies in ageing and neurodegenerative disorders
New in @NatureComms: I'm thrilled to share with you our latest work that applies #srRNAseq to understand splicing accuracy across human introns, tissues and in the context of ageing and neurodegeneration https://t.co/KapUzy4OQP
— SoniaRuiz (@sonigruiz) January 27, 2025
Congratulations to @soniagr.bsky.social, rytenlab.com team, et al for this project re-analyzing #GTEx human gene expression data uniformly re-processed with #recount3
It was a pleasure working with Sonia G & Mina Ryten
doi.org/10.1038/s414…
@bioconductor.bsky.social @lieberinstitute.bsky.social
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor.bsky.social) January 28, 2025 at 3:54 PM
[image or embed]