Benchmark of cellular deconvolution methods using a multi-assay dataset from postmortem human prefrontal cortex
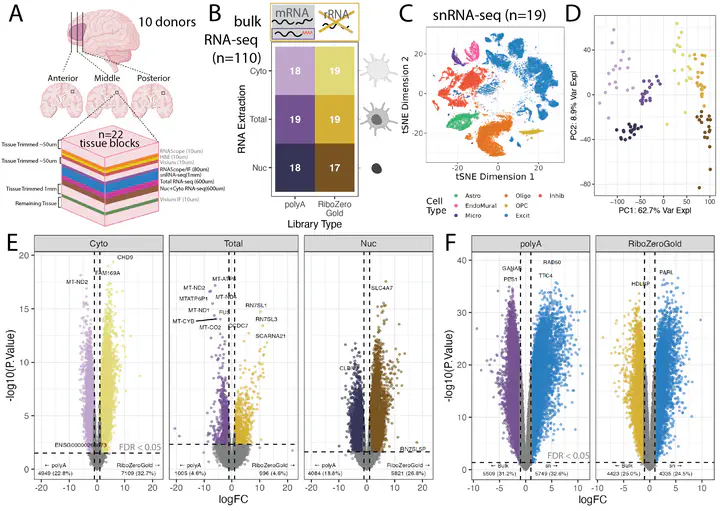 Image credit: bioRxiv
Image credit: bioRxiv
Abstract
Cellular deconvolution of bulk RNA-sequencing data using single cell/nuclei RNA-seq reference data is an important strategy for estimating cell type composition in heterogeneous tissues, such as the human brain. Here, we generate a multi-assay dataset in postmortem human dorsolateral prefrontal cortex from 22 tissue blocks, including bulk RNA-seq, reference snRNA-seq, and orthogonal measurement of cell type proportions with RNAScope/ImmunoFluorescence. We use this dataset to evaluate six deconvolution algorithms. Bisque and hspe were the most accurate methods. The dataset, as well as the Mean Ratio gene marker finding method, is made available in the DeconvoBuddies R/Bioconductor package.
Hey #deconvolution fans! 👀 We’ve got an exciting new publication for you: a benchmark of 6 popular deconvolution methods on a multimodal human brain #DLPFC dataset 🧠🧬 Now out in Genome Biology: doi.org/10.1186/s130...
@lieberinstitute.bsky.social @jhubiostat.bsky.social
— Louise Huuki-Myers (@lahuuki.bsky.social) April 11, 2025 at 10:35 AM
[image or embed]
Excited to announce that our #deconvolution benchmark has now been published in @GenomeBiology ! 📊
— Louise Huuki-Myers (@lahuuki) April 9, 2025
📄 https://t.co/A3oQERcuqF
Check out my blog post for a high level overview: https://t.co/WtyplwOi0e https://t.co/zeDFrUPLKa