Data-driven Identification of Total RNA Expression Genes (TREGs) for Estimation of RNA Abundance in Heterogeneous Cell Types
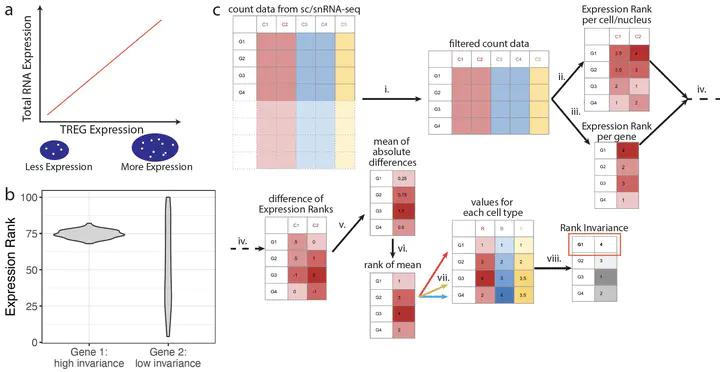 Image credit: bioRxiv
Image credit: bioRxiv
Abstract
Next-generation sequencing technologies have facilitated data-driven identification of gene sets with different features including genes with stable expression, cell-type specific expression, or spatially variable expression. Here, we aimed to define and identify a new class of "control" genes called Total RNA Expression Genes (TREGs), which correlate with total RNA abundance in heterogeneous cell types of different sizes and transcriptional activity. We provide a data-driven method to identify TREGs from single cell RNA-sequencing (RNA-seq) data, available as an R/Bioconductor package at https://bioconductor.org/packages/TREG. We demonstrated the utility of our method in the postmortem human brain using multiplex single molecule fluorescent in situ hybridization (smFISH) and compared candidate TREGs against classic housekeeping genes. We identified AKT3 as a top TREG across five brain regions, especially in the dorsolateral prefrontal cortex.
Excited to share my first 🥇 first-author manuscript: "Data-driven Identification of Total RNA Expression Genes (TREGs) for Estimation of RNA Abundance in Heterogeneous Cell Types" 👩🔬 @LieberInstitute #scitwitter
— Louise Huuki-Myers (@lahuuki) May 3, 2022
Now a @biorxivpreprint ! 🎉
📰 https://t.co/OJuAWndW51 pic.twitter.com/VZ8xzJpcyv
To end the week: CONGRATS @lahuuki for your first first-author @biorxivpreprint 📜! 🎉 The first of many I'm 💯 sure!#rstats @LieberInstitute
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) April 30, 2022
📦@Bioconductor https://t.co/5f79NtYyFu
💻@GitHub https://t.co/Hpg8rKLsst
📔#pkgdown https://t.co/6of1LCtxgC
👀https://t.co/J0uJA15liE pic.twitter.com/uUPZEUXns9