Profiling gene expression in the human dentate gyrus granule cell layer reveals insights into schizophrenia and its genetic risk
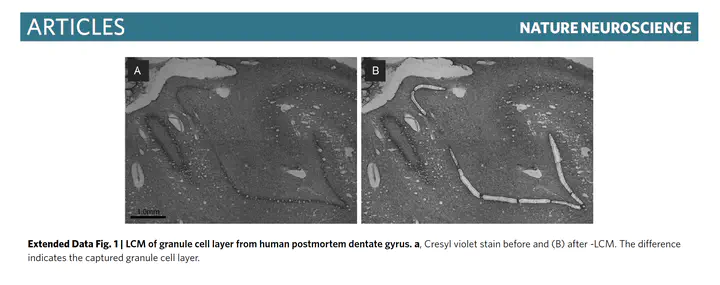 Image credit: Nature Neuroscience
Image credit: Nature Neuroscience
Abstract
Specific cell populations may have unique contributions to schizophrenia but may be missed in studies of homogenate tissue. Here laser capture microdissection followed by RNA sequencing (LCM-seq) was used to transcriptomically profile the granule cell layer of the dentate gyrus (DG-GCL) in human hippocampus and contrast these data to those obtained from bulk hippocampal homogenate. We identified widespread cell-type-enriched aging and genetic effects in the DG-GCL that were either absent or directionally discordant in bulk hippocampus data. Of the ~9 million expression quantitative trait loci identified in the DG-GCL, 15% were not detected in bulk hippocampus, including 15 schizophrenia risk variants. We created transcriptome-wide association study genetic weights from the DG-GCL, which identified many schizophrenia-associated genetic signals not found in transcriptome-wide association studies from bulk hippocampus, including GRM3 and CACNA1C. These results highlight the improved biological resolution provided by targeted sampling strategies like LCM and complement homogenate and singlenucleus approaches in human brain.
while we're all dealing with this COVID19 outbreak right now, I'm happy to share our latest work on identifying expression quantitative trait loci (eQTLs) and other molecular associations (age + psychiatric diagnoses) in an important cell population in the human brain (1/n) https://t.co/FJQcnzvvE7
— Andrew Jaffe (@andrewejaffe) March 17, 2020