Incomplete annotation has a disproportionate impact on our understanding of Mendelian and complex neurogenetic disorders
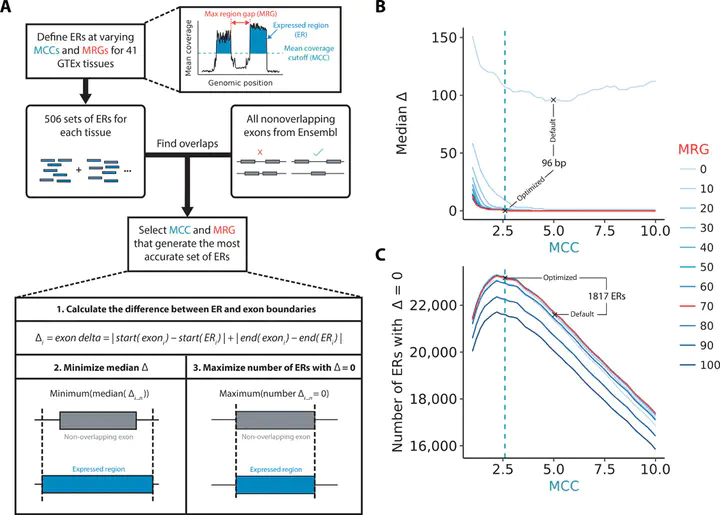 Image credit: Science Advances
Image credit: Science Advances
Abstract
Growing evidence suggests that human gene annotation remains incomplete; however, it is unclear how this affects different tissues and our understanding of different disorders. Here, we detect previously unannotated transcription from Genotype-Tissue Expression RNA sequencing data across 41 human tissues. We connect this unannotated transcription to known genes, confirming that human gene annotation remains incomplete, even among well-studied genes including 63% of the Online Mendelian Inheritance in Man–morbid catalog and 317 neurodegeneration-associated genes. We find the greatest abundance of unannotated transcription in brain and genes highly expressed in brain are more likely to be reannotated. We explore examples of reannotated disease genes, such as SNCA, for which we experimentally validate a previously unidentified, brain-specific, potentially protein-coding exon. We release all tissue-specific transcriptomes through vizER: http://rytenlab.com/browser/app/vizER. We anticipate that this resource will facilitate more accurate genetic analysis, with the greatest impact on our understanding of Mendelian and complex neurogenetic disorders.
Occasionally I encounter researchers, who suggest bulk RNA-seq transcriptomics is "sort" of solved, human annotation is "sort" of complete, and it's probably not worth spending long hours in looking into novel isoforms, splicing events: For them https://t.co/6NBYOz9OZP 1/2 pic.twitter.com/9LMAOo24KA
— Hirak Sarkar @hirak@genomic.social (@hrksrkr) June 12, 2020
Welcome to #academic twitter David Zhang! 🙌🏽🥳
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) February 5, 2019
He’s an awesome researcher and excellent at translating ideas into #rstats code as well as beautiful figures! 🤩
@DavidZh03027445
He is the co-lead author of our recent preprint https://t.co/VVr9Qfm6R7
I bet many PIs will 👀 him!