SpatialExperiment: infrastructure for spatially resolved transcriptomics data in R using Bioconductor
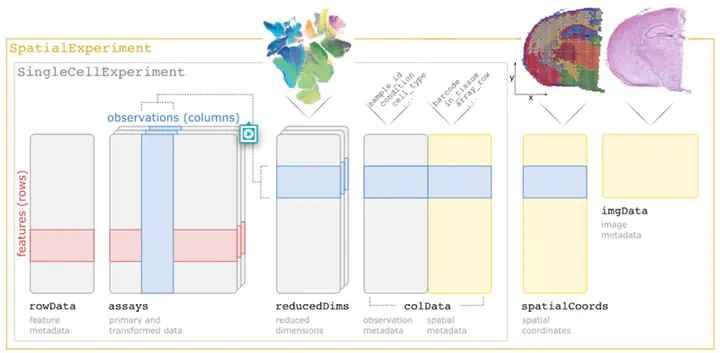 Image credit: bioRxiv
Image credit: bioRxiv
Abstract
Summary SpatialExperiment is a new data infrastructure for storing and accessing spatially resolved transcriptomics data, implemented within the R/Bioconductor framework, which provides advantages of modularity, interoperability, standardized operations, and comprehensive documentation. Here, we demonstrate the structure and user interface with examples from the 10x Genomics Visium and seqFISH platforms, and provide access to example datasets and visualization tools in the STexampleData, TENxVisiumData, and ggspavis packages. Availability and Implementation The SpatialExperiment, STexampleData, TENxVisiumData, and ggspavis packages are available from Bioconductor. The package versions described in this manuscript are available in Bioconductor version 3.15 onwards. Supplementary information Supplementary tables and figures are available at Bioinformatics online.
Are you analyzing spatially-resolved transcriptomics data? Visium from @10xGenomics? You might want to use #SpatialExperiment which provides a common infrastructure fully compatible with @Bioconductor's ecosystem #rstats
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) April 29, 2022
More details? Open access paper 📜https://t.co/4Y2RN4Wz2z https://t.co/49wE5Dv5HF pic.twitter.com/QpI4lvCF2r
Are you working with spatial transcriptomics data such as Visium from @10xGenomics? Then you'll be interested in #SpatialExperiment 📦 led by @drighelli @lmwebr @CrowellHL with contributions by @PardoBree @shazanfar A Lun @stephaniehicks @drisso1893 🌟
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) January 29, 2021
📜 https://t.co/r36qlakRJe pic.twitter.com/cWIiwLFitV