Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex
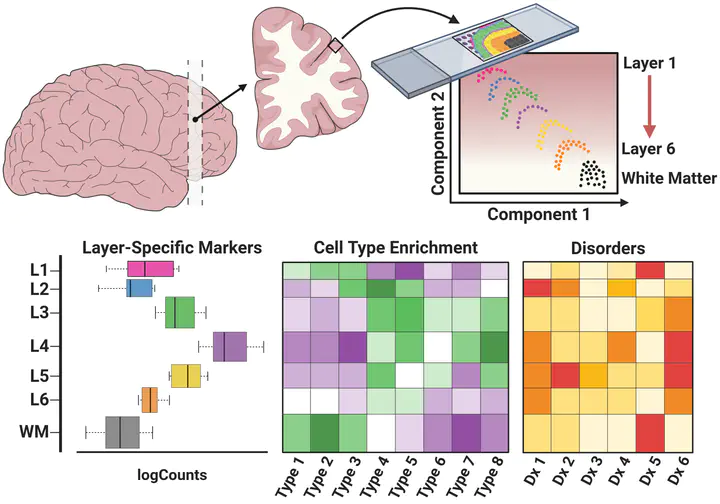 Image credit: bioRxiv
Image credit: bioRxiv
Abstract
We used the 10x Genomics Visium platform to define the spatial topography of gene expression in the six-layered human dorsolateral prefrontal cortex. We identified extensive layer-enriched expression signatures and refined associations to previous laminar markers. We overlaid our laminar expression signatures on large-scale single nucleus RNA-sequencing data, enhancing spatial annotation of expression-driven clusters. By integrating neuropsychiatric disorder gene sets, we showed differential layer-enriched expression of genes associated with schizophrenia and autism spectrum disorder, highlighting the clinical relevance of spatially defined expression. We then developed a data-driven framework to define unsupervised clusters in spatial transcriptomics data, which can be applied to other tissues or brain regions in which morphological architecture is not as well defined as cortical laminae. Last, we created a web application for the scientific community to explore these raw and summarized data to augment ongoing neuroscience and spatial transcriptomics research (http://research.libd.org/spatialLIBD).
pleased to announce that this paper is now out in @NatureNeuro : https://t.co/m5L1YvETzN ! check out full text here: https://t.co/zMdMTA5QG4 . congrats to @kr_maynard @lcolladotor @lmwebr @stephaniehicks and @martinowk and other investigators at @LieberInstitute & @10xGenomics https://t.co/StYedlFMHc
— Andrew Jaffe (@andrewejaffe) February 8, 2021
🔥off the press! 👀 our @biorxivpreprint on human 🧠brain @LieberInstitute spatial 🌌🔬transcriptomics data 🧬using Visium @10xGenomics🎉#spatialLIBD
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) February 29, 2020
🔍https://t.co/RTW0VscUKR
👩🏾💻https://t.co/bsg04XKONr
📚https://t.co/FJDOOzrAJ6
📦https://t.co/Au5jwADGhYhttps://t.co/PiWEDN9q2N pic.twitter.com/aWy0yLlR50