Intro RNA-seq LCG-UNAM 2026
Overview
Here you can find the files for the February 2026 introduction to R, RStudio and RNA-sequencing (RNA-seq) course for LCG-UNAM at CCG-UNAM (February 10-13, 2026). The rest of the chapters will be in Spanish.
Instructor:
Teaching assistants:
Guest presenters:
- Melissa Mayén Quiroz, Bluesky
- Maria Gutiérrez-Arcelus, Bluesky
- Gabriel Ramírez-Vilchis, Bluesky
- Daianna González Padilla, Bluesky
Course Coordinators:
- Heladia Salgado
- Julio Collado Vides
Download the materials for this course with usethis::use_course('lcolladotor/rnaseq_LCG-UNAM_2026') or view online at lcolladotor.github.io/rnaseq_LCG-UNAM_2026.
Code of Conduct
We’ll follow the CDSB code of conduct comunidadbioinfo.github.io/codigo-de-conducta/ as well as version 1.4.0 of the Bioconductor code of conduct bioconductor.github.io/bioc_coc_multilingual/.
For reporting any violations of the code of conduct, report them to the Instructor and/or Course Coordinators.
Course Schedule
Local times in Cuernavaca, Mexico
- Tuesday February 10:
- Wednesday February 11:
- 9-11 am: introduction to Bioconductor
- 12-2 pm: expression data R/Bioconductor objects and visualization: SummarizedExperiment and iSEE
- 12-1 pm: SummarizedExperiment intro
- 1-2 pm: Melissa Mayén Quiroz will teach you about iSEE and showcase an iSEE app she made for LieberInstitute/Hb_VTA_projector_profiling.
- Thursday February 12:
- 9-11 am: making your own website with postcards by Gabriel Ramírez-Vilchis
- 12-2 pm: downloading RNA-seq data using recount3
- Friday February 13:
- 9-11 am: exploring statistical models with ExploreModelMatrix
- 12-1:30 pm: current examples of RNA-seq data analysis
- Guest presentation by Maria Gutiérrez-Arcelus
- Guest presentation by Daianna González Padilla
- Guest presentation by Gabriel Ramírez-Vilchis
- 1:30-2 pm: differential expression analysis with limma
- review
- R/Bioconductor-powered Team Data Science overview
- Potentially after 2 pm: open discussion (career paths, networking, etc)
External links
- Cursos LCG
- GitHub source code
- Zoom (ask)
- LCG-UNAM RStudio server
- CDSB
- Slack CDSB Mexico, in particular the
intro_rnaseq_lcg_2026channel.- You can join this Slack workspace through this link.
- Bioconductor Zulip community
- LieberInstitute/template_project
- Example real project by Daianna González Padilla: LieberInstitute/smoking-nicotine-mouse
Course Prerequisites
- Install R 4.5.x from CRAN.
- Install Positron which will be our Interactive Development Environment (IDE) of choice for the course.
- Alternatively, you could install RStudio IDE version RStudio 2026.01.0 or newer.
- In either case, you will also need to install Air for automatic code formatting.
- You will also have to requesst access to the GitHub Education pack in order to use GitHub Copilot Pro 🤖.
- Finally install the following R packages:
## For installing Bioconductor packages
if (!requireNamespace("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
## Install required packages
BiocManager::install(
c(
"usethis", ## Utilities
"here",
"biocthis",
"lobstr",
"postcards",
"sessioninfo",
"SummarizedExperiment", ## Main containers / vis
"iSEE",
"edgeR", ## RNA-seq
"ExploreModelMatrix",
"limma",
"recount3",
"pheatmap", ## Visualization
"ggplot2",
"patchwork",
"RColorBrewer",
"ComplexHeatmap",
"spatialLIBD" ## Advanced
)
)Getting set up
Here are more step by step instructions for the course prerequisites.
You must install R and Positron (available from Posit) on your computing environment. As an alternative to Positron, you can install RStudio IDE.
These are three different applications that must be installed separately before they can be used together:
Ris the core underlying programming language and computing engine that we will be learning in this courseRStudio IDEis an interface into R that makes many aspects of using and programming R simplerPositronis a new interface for working with R developed by Posit (formerly RStudio). All new features are being implemented inPositron, but as it was just recently released, it might have some bugs. If you are choosing betweenRStudio IDEandPositron, check https://positron.posit.co/faqs.html#is-rstudio-going-away.
R, RStudio IDE, and Positron are available for Windows, macOS, and most flavors of Unix and Linux. Please download the version that is suitable for your computing setup.
Throughout the course, we will make use of numerous R add-on packages that must be installed over the Internet. Packages can be installed using the install.packages() function in R. For example, to install the tidyverse package, you can run
in the R console.
We will also use Git to version control your code, which you can install following the Happy Git and GitHub for the useR book. Through the GitHub Education pack we will get free access to GittHub Copilot Pro to help us write code: think about it as an enhanced auto-complete. Finally, we’ll use Air for automatically formatting our R code.
How to Download R for Windows
Go to https://cran.r-project.org and
Click the link to “Download R for Windows”
Click on “base”
Click on “Download R 4.5.2 for Windows”
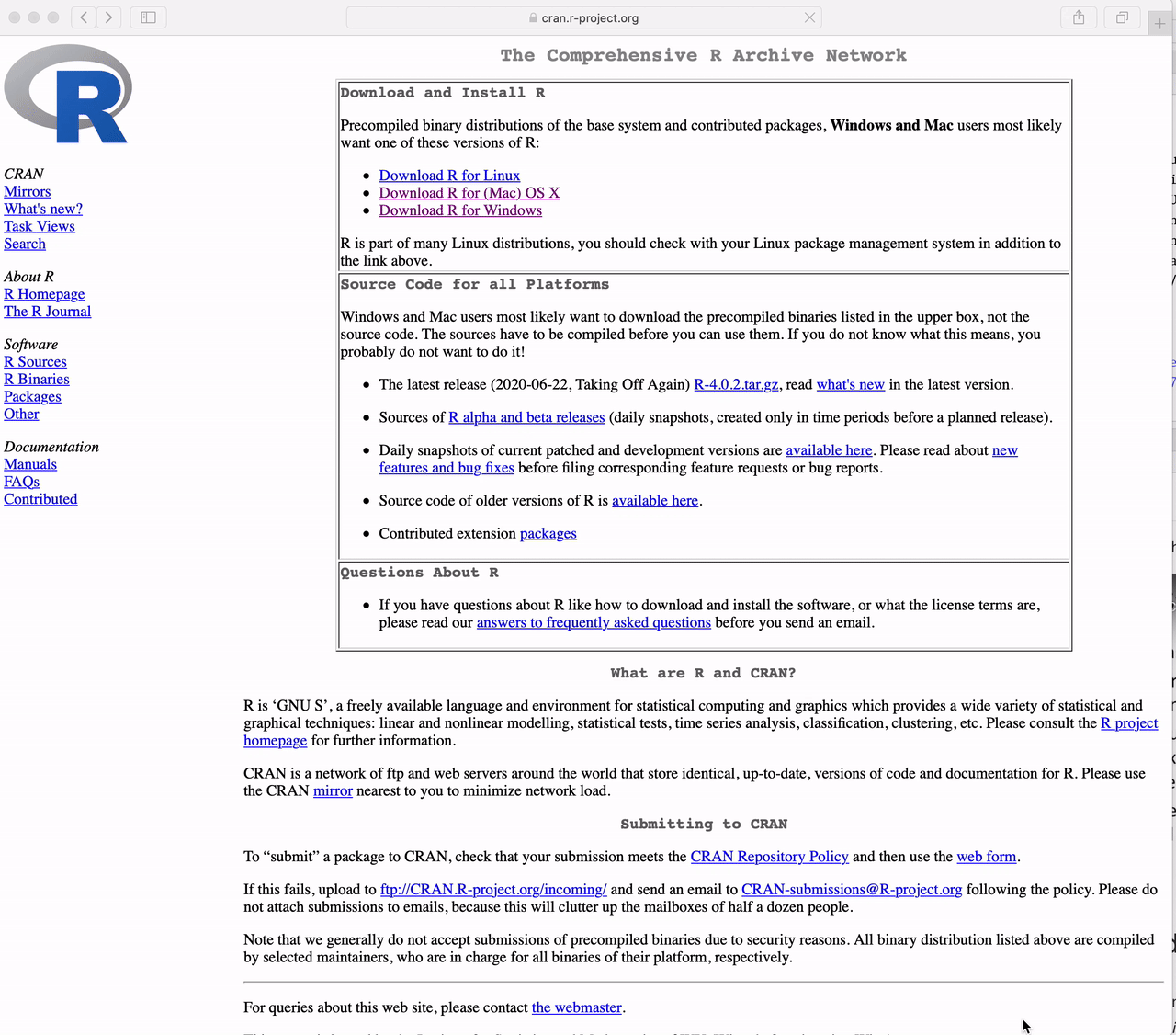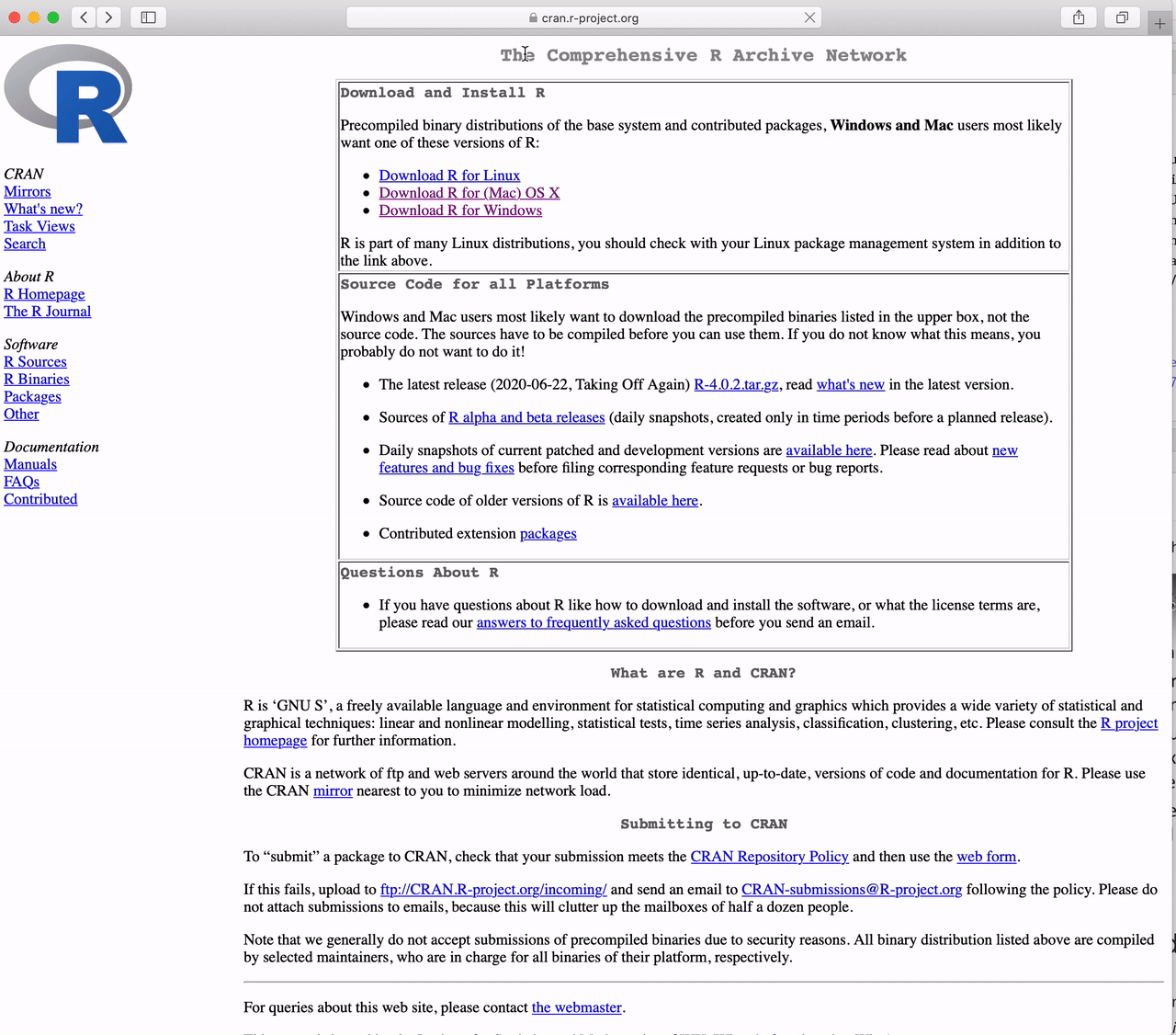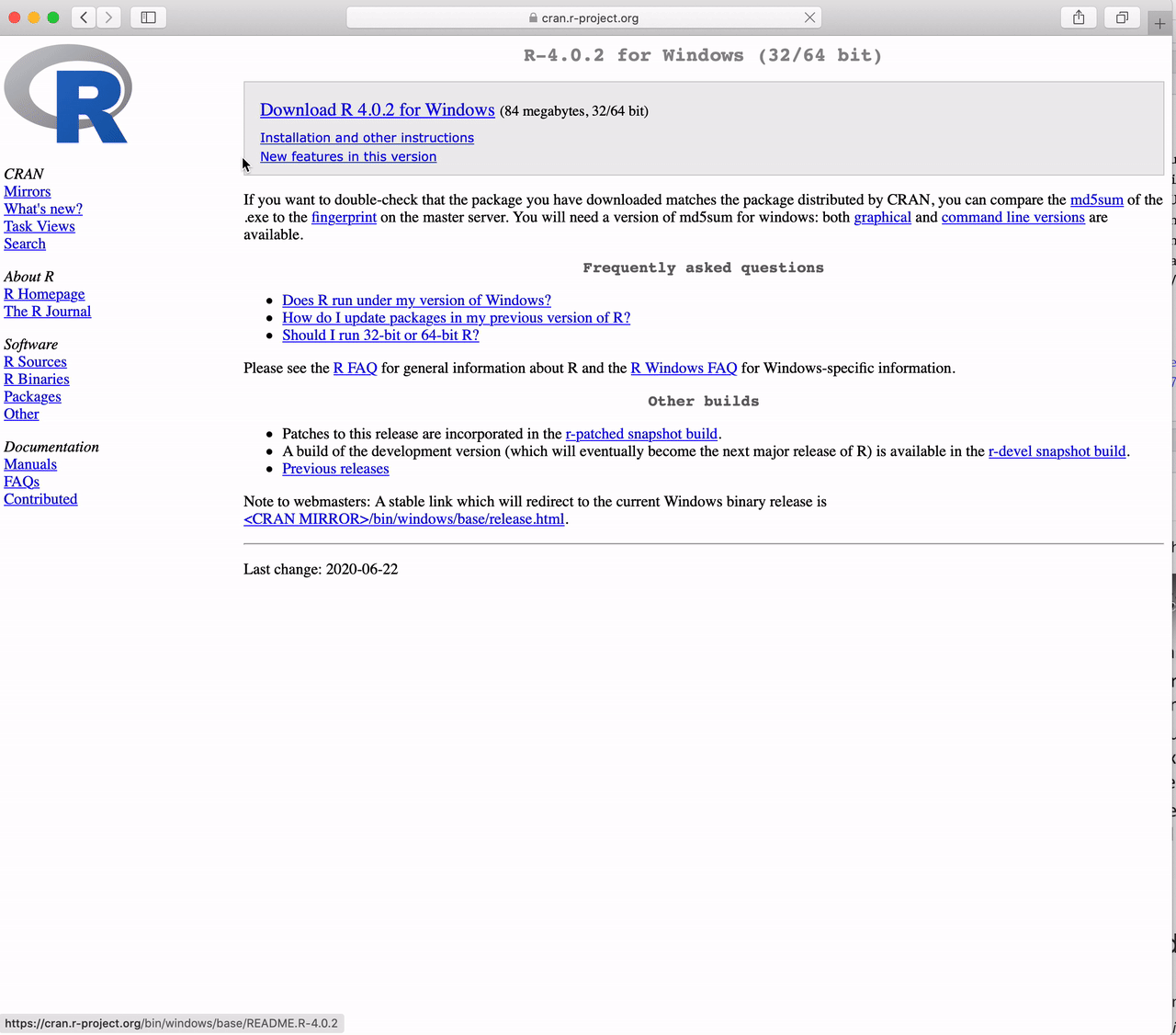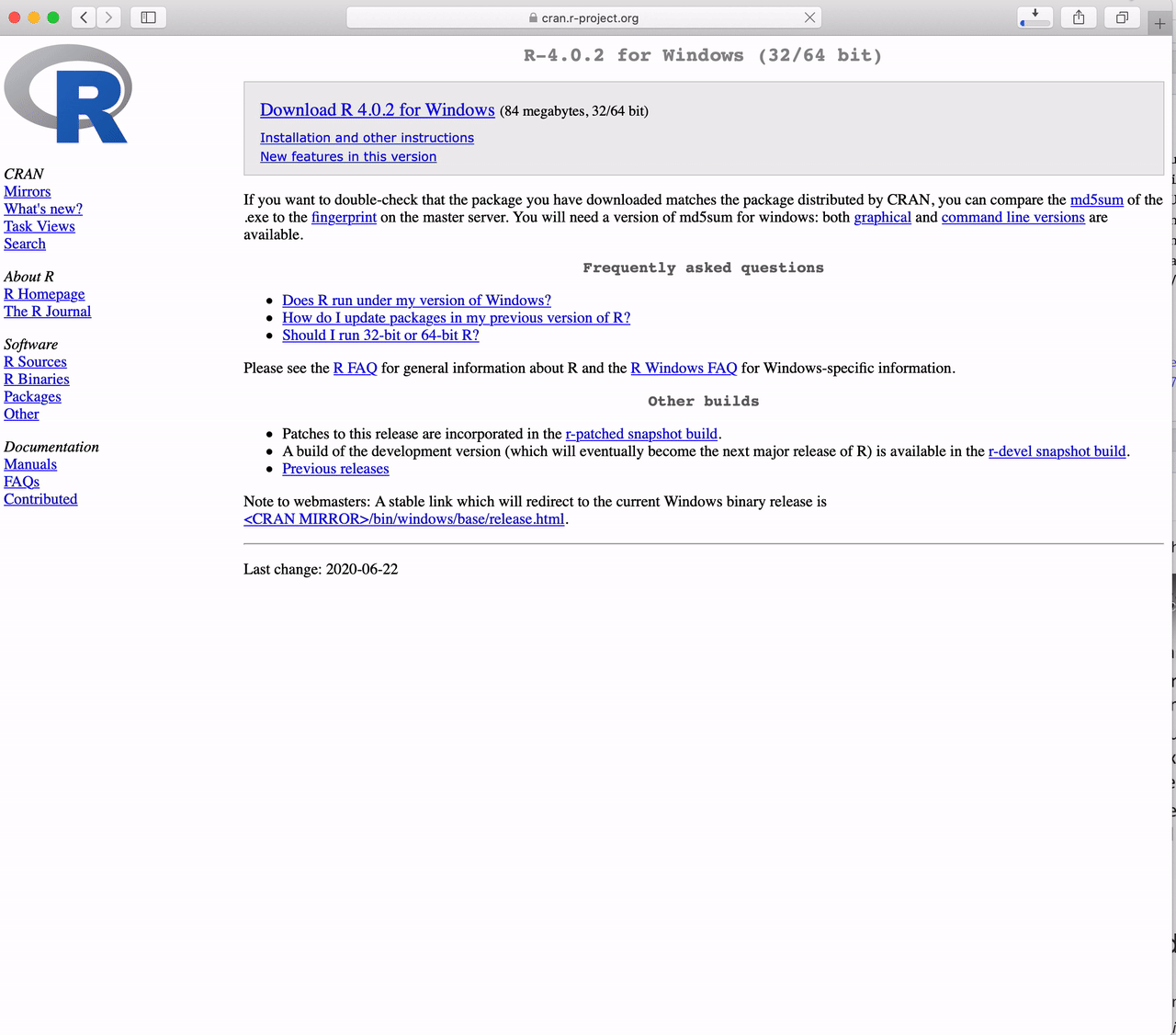
The version in the video is not the latest version of R. Please download the latest version.
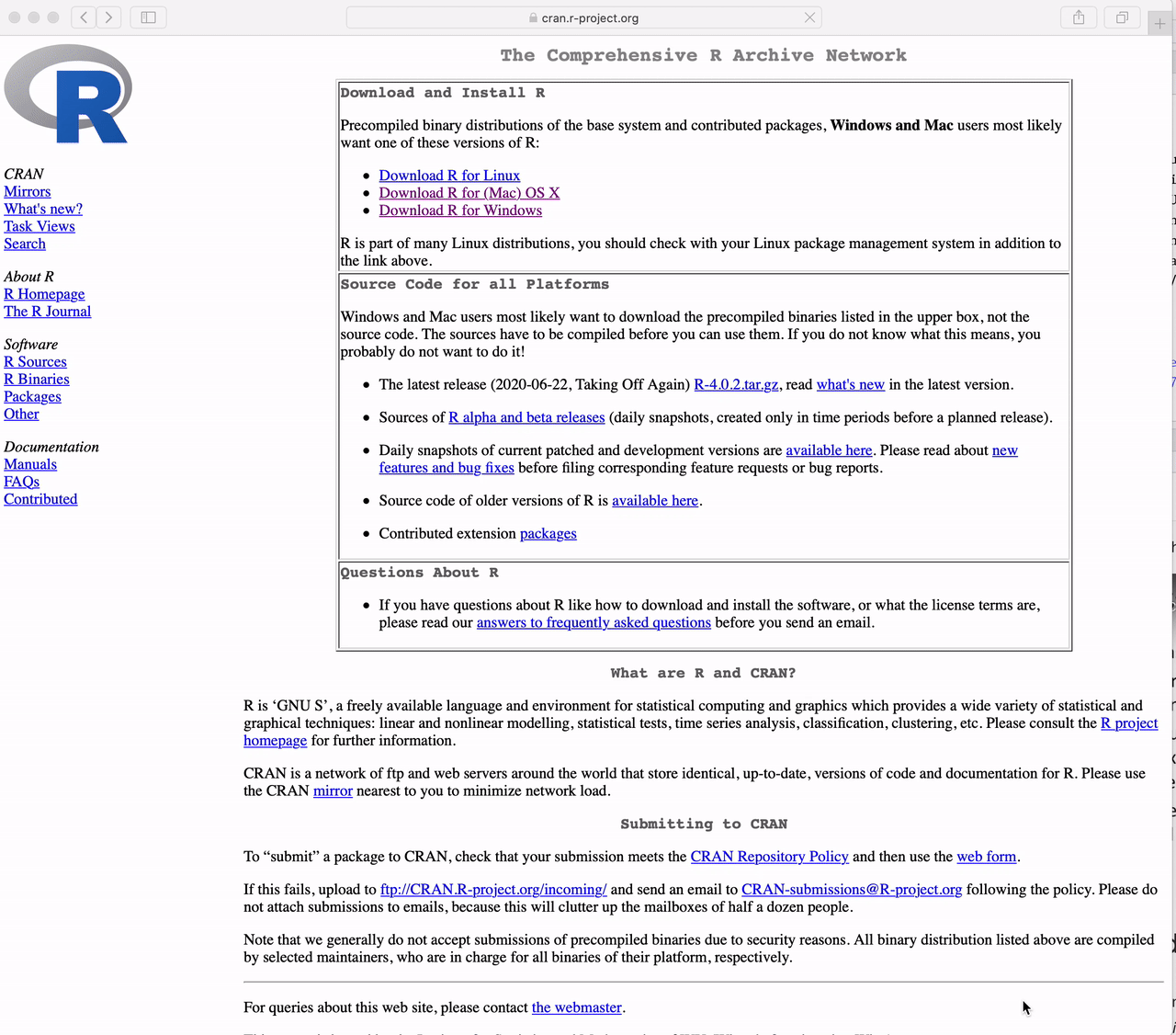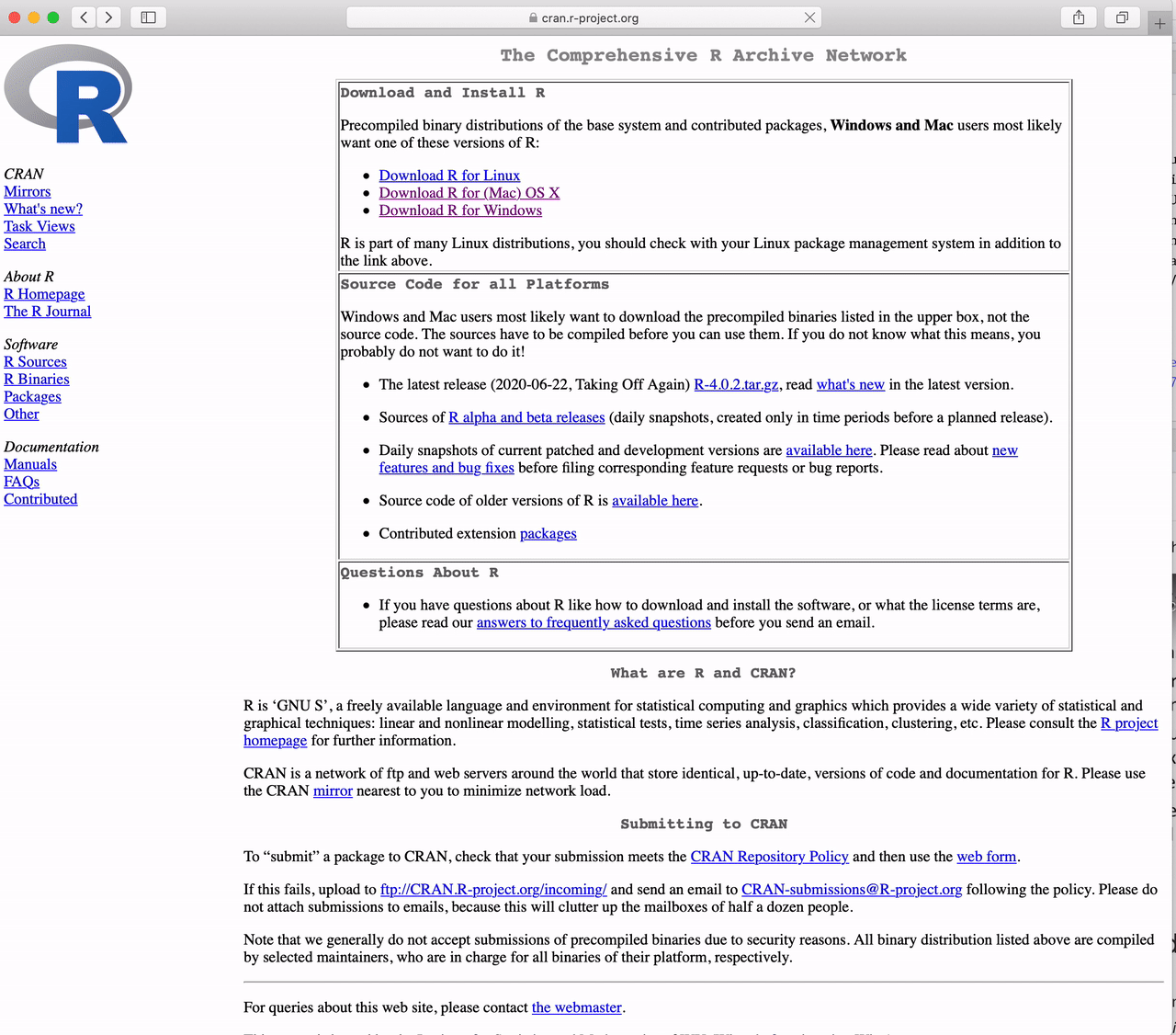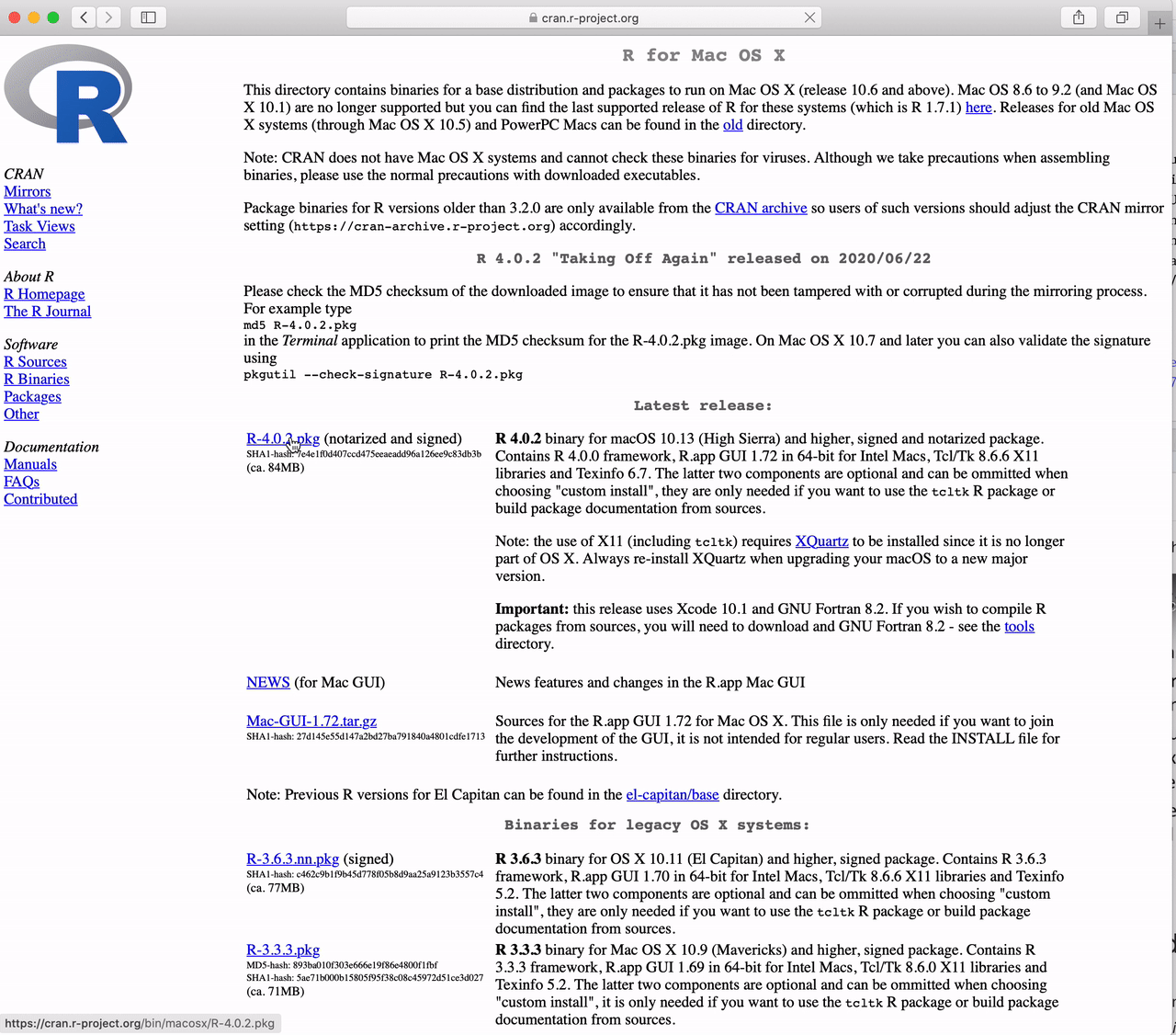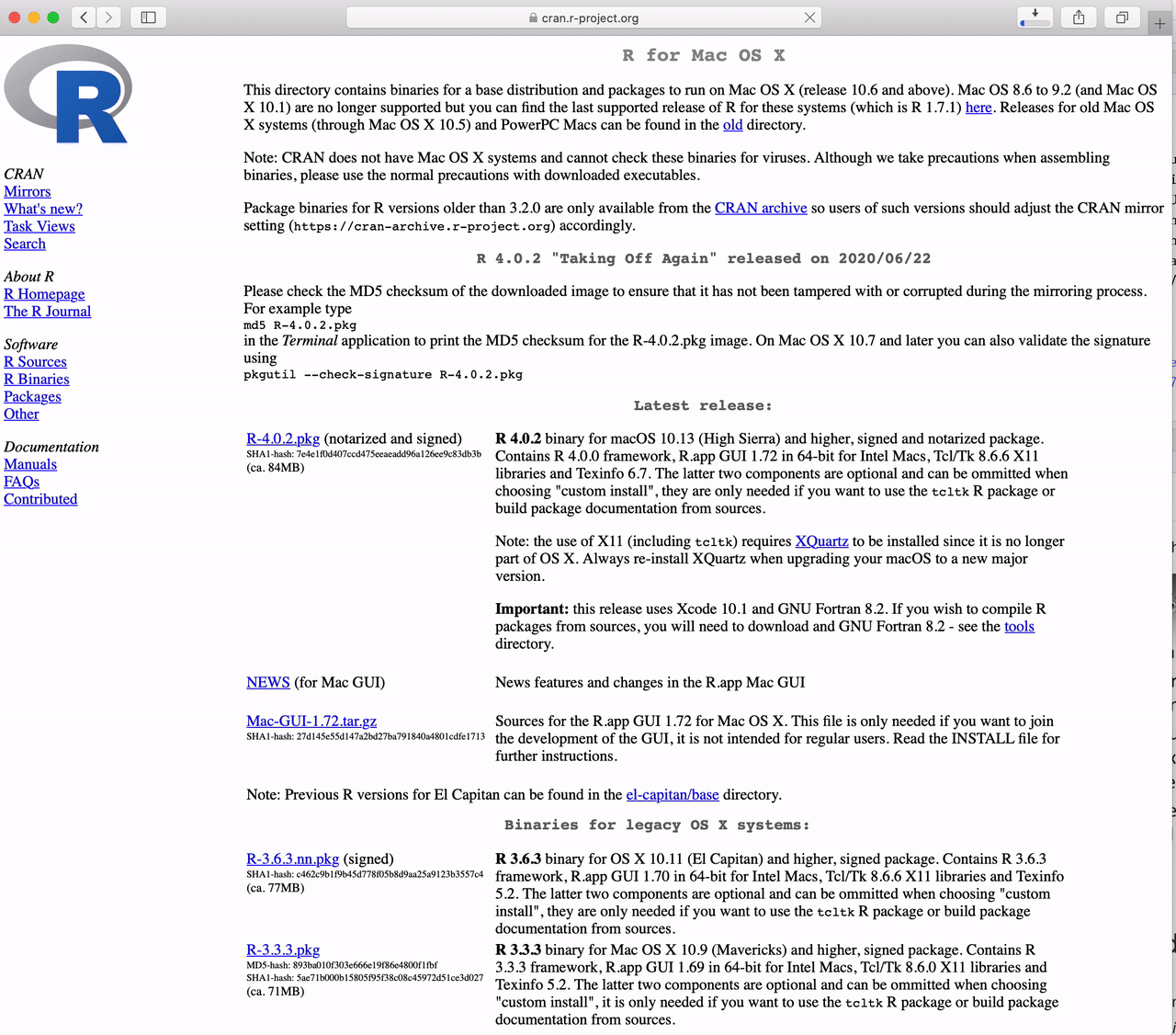
How to Download R for the Mac
Go to https://cran.r-project.org and
Click the link to “Download R for (Mac) OS X”.
Click on “R-4.5.2-arm64.pkg” (for Apple Silicon Macs)
The version in the video is not the latest version of R. Please download the latest version.
How to Download RStudio IDE
Go to https://posit.co/ (formerly https://rstudio.com) and
Click on “Free & Open Source” in the top menu
Click on “RStudio IDE” in the drop down menu
Click the button that says “DOWNLOAD RSTUDIO DESKTOP”
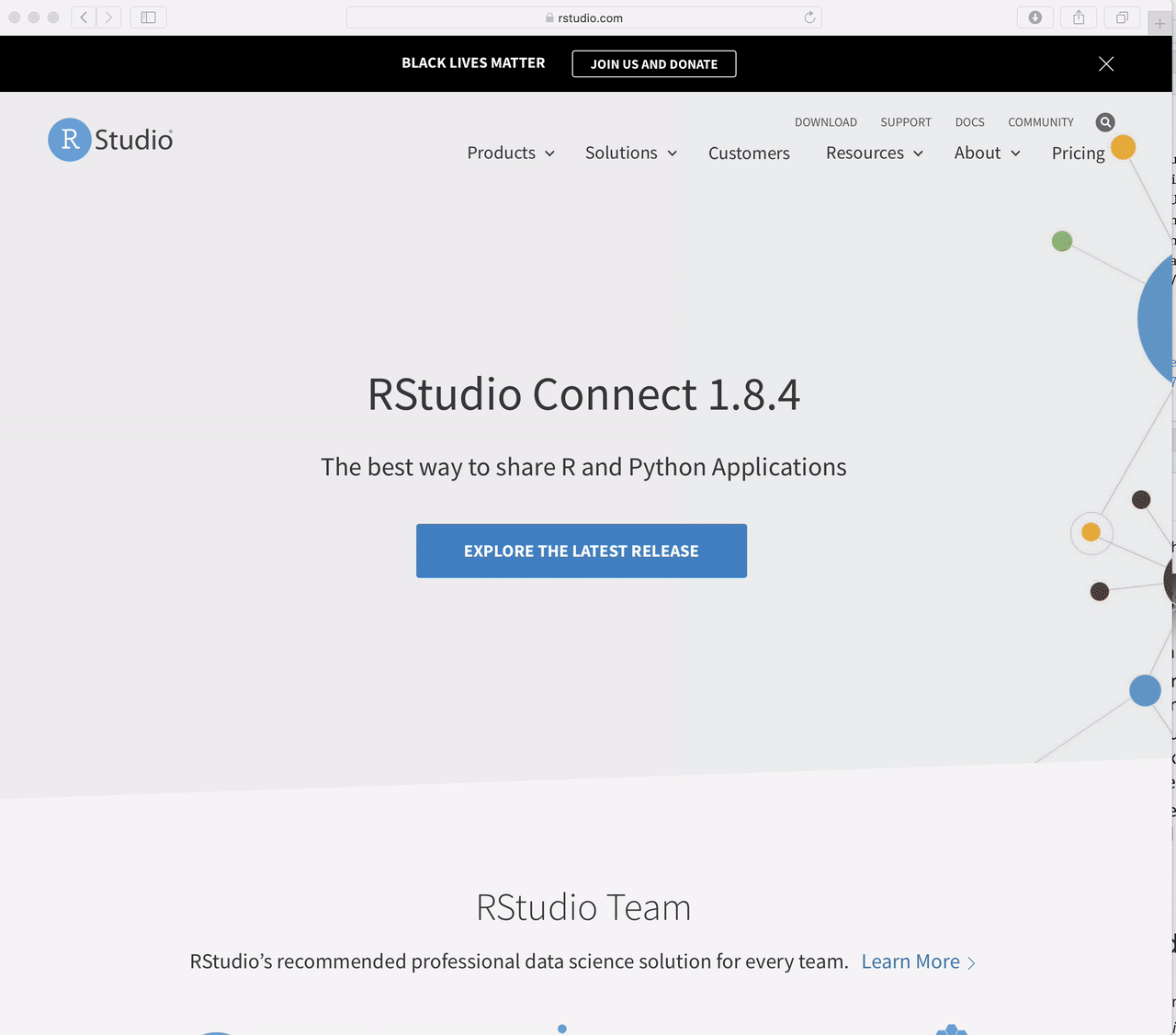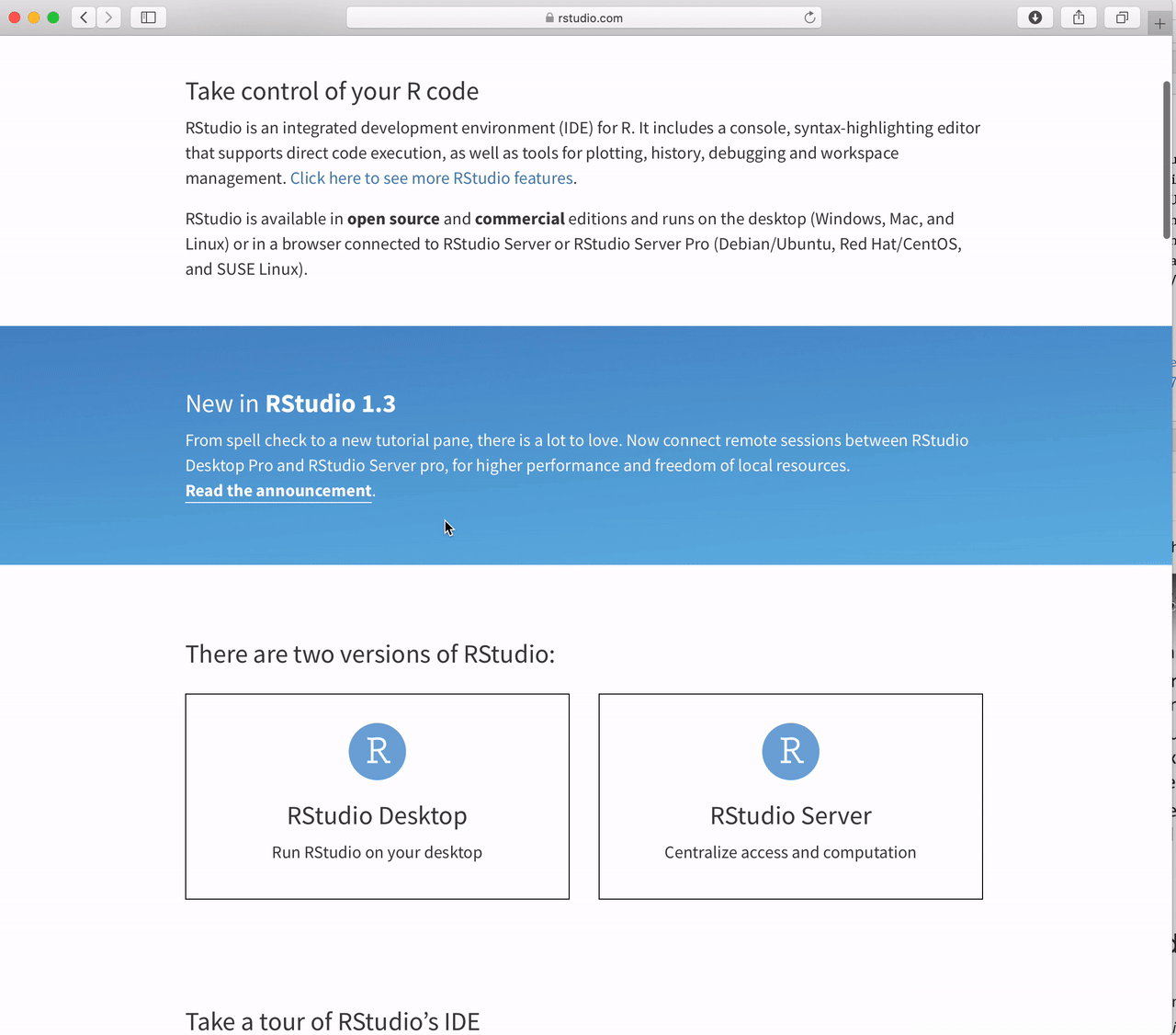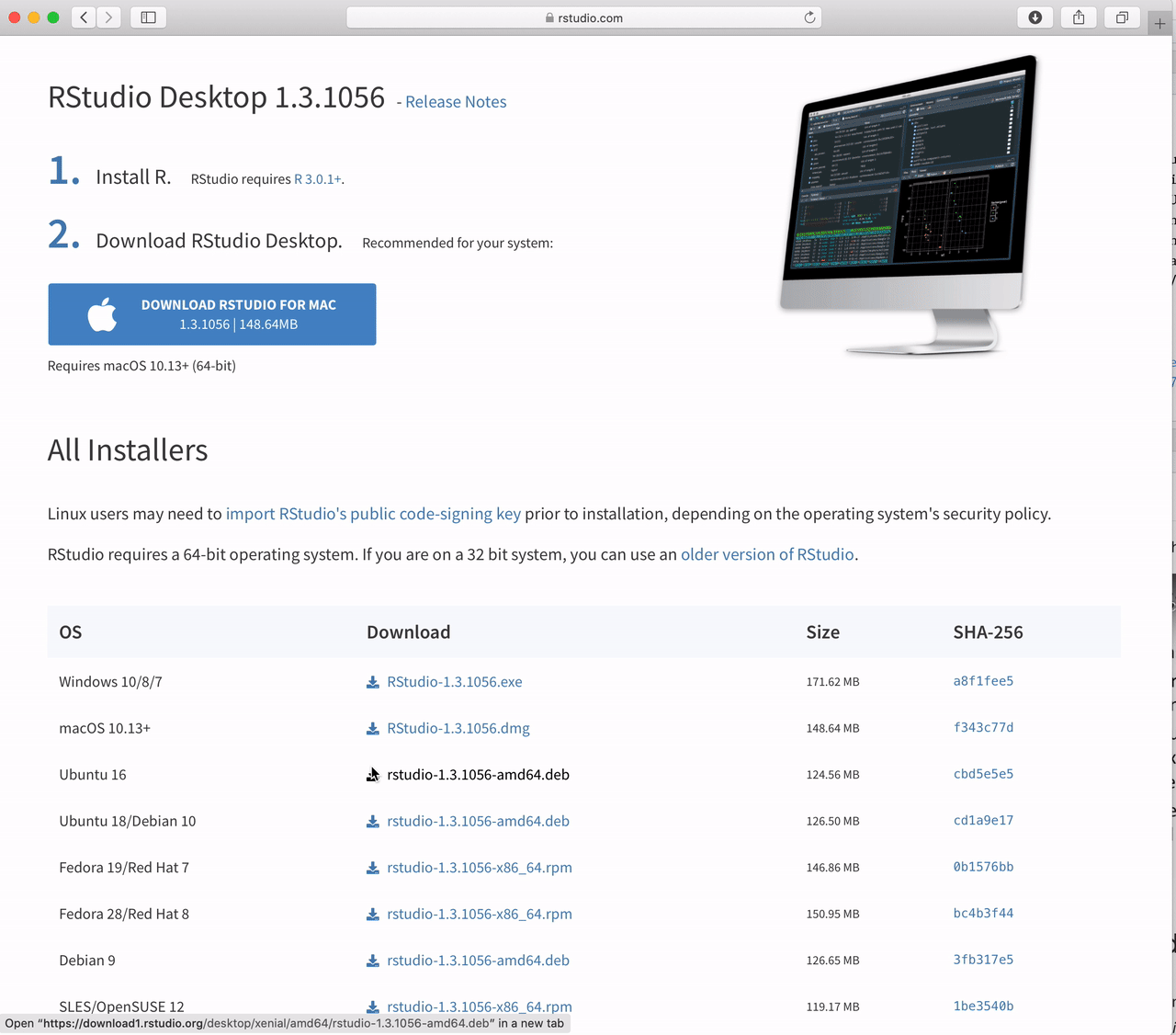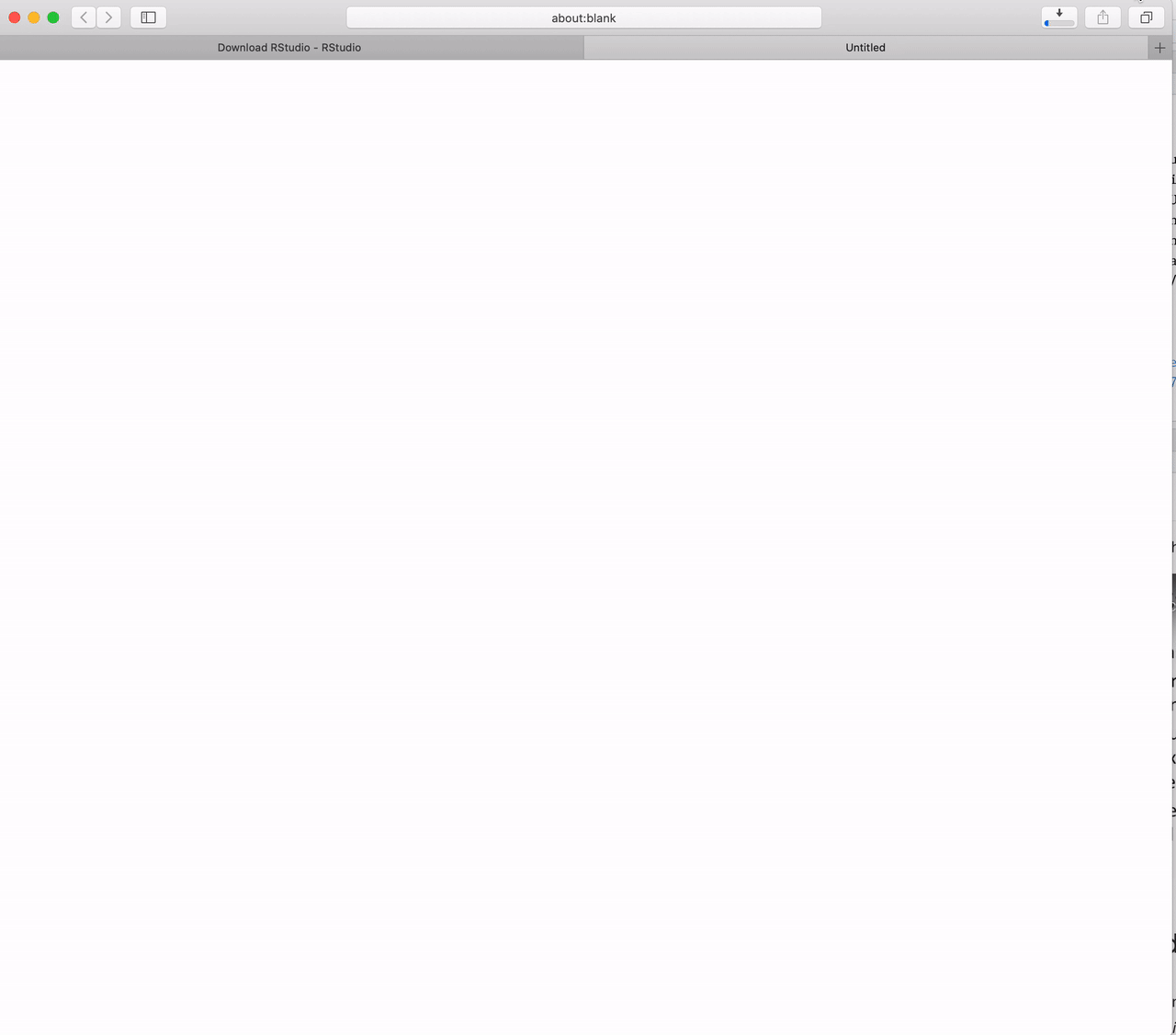
Click the button under “Download RStudio” which will take you to https://posit.co/download/rstudio-desktop/
Under the section “All Installers and Tarballs” choose the file that is appropriate for your operating system.
NOTE: The video shows how to download RStudio for the Mac but you should download RStudio for whatever computing setup you have.
How to Download Positron
Go to https://posit.co/ and
Click on “Free & Open Source” in the top menu
Click on “Positron” in the drop down menu
Click on “Download Positron” which will take you to https://positron.posit.co/download.html
Read the license agreement.
Accept the license agreement.
On the table with all the download options, choose the file that is appropriate for your operating system.
How to Download Git
Install Git on your computer following the detailed instructions at https://happygitwithr.com/install-git, which will depend on your operating system.
Create your GitHub account
Create a personal GitHub account following the instructions at https://docs.github.com/en/get-started/signing-up-for-github/signing-up-for-a-new-github-account.
Following Jeff Leek’s advice on How to be a modern scientist that was part of my team’s bootcamps at https://lcolladotor.github.io/bioc_team_ds/how-to-be-a-modern-scientist.html, try to choose a username that will be the same one you use on your email and other work-related social media platforms such as Bluesky, LinkedIn, etc.
If you are using Positron, under the “Accounts” options (bottom left corner), click on “Sign in with GitHub \[...\]”, provide the 8 digit code, and authorize “posit-dev”.
How to Apply for GitHub Education and configure GitHub Copilot
Go to https://github.com/education and
- Click on “Join GitHub education” (likely https://github.com/settings/education/benefits?locale=en-US)
- Sign in to your GitHub account if you haven’t logged in
- Click on “Start Application”
- There will be a few options to verify that you are a student. One of them is by having a photo of your student ID with dates showing that your student ID is still valid. I’ve used a screenshot of my Faculty website at https://publichealth.jhu.edu/faculty/4647/leonardo-collado-torres when validating my information.
- Wait a few days for GitHub to process your application.
- Once your application has been approved, if you are using
RStudio IDEfollow the instructions at https://docs.posit.co/ide/user/ide/guide/tools/copilot.html to enable GitHub Copilot in RStudio IDE. I have the “Index project files with GitHub Copilot” checkbox enabled on my preferences. - If you are using
Positron, follow the instructions at https://positron.posit.co/assistant to enable “Positron assistant”. Then under the “Positron Assistant” chat, click on “Add Anthropic as a Chat Provider”, then select “GitHub Copilot” and click “Sign in”.
How to get Air (auto-code formatter)
Go to https://posit-dev.github.io/air/ and
Click on “RStudio” if you are using
RStudio IDE. That will take you to https://posit-dev.github.io/air/editor-rstudio.html where you will find the detailed installation instructions.- Note that I recommend enabling the
Tools -> Global Options -> Code -> Savingand checkReformat documents on saveoption. Particularly if you are working on your own code.
- Note that I recommend enabling the
Click on “VS Code and Positron” if you are using
Positron. That will take you to https://posit-dev.github.io/air/editor-vscode.html where some options and configuration steps are described. Note however, thatAiris already available fromPositron. You will need to open yoursettings.jsonfile from the Command Palette (Shift + Cmd + Pon Mac/Linux,Shift + Ctrl + Pon Windows):Run
Preferences: Open Workspace Settings (JSON)to modify workspace specific settings (recommended).Run
Preferences: Open User Settings (JSON)to modify global user settings (preferred option if you are not working with code from others). Then edit yoursettings.jsonwith the relevant lines about R code formatting andAiras shown below:
- You can also configure a few options at positron://settings/air.dependencyLogLevels and related settings.
Course Evaluation
- Participation
- (main evidence) written and/or verbal participation during class
- This is tracked by day by the TA. Each day counts as a category.
- (optional, strongly encouraged for your own career) Community building: CDSB Slack (introduce yourself), Bluesky, GitHub, etc. These are tracked by the TA: please let the TA know of your participations.
- GitHub issues on the course repository
- CDSB Slack #bienvenida channel
- Bioconductor Zulip #introductions channel
- Bluesky posts related to the course
- Grading:
- 0%: no participation
- 25%: participation in only 1 out of 8 categories (4 days, 4 public participation categories)
- 50%: participation in 2 out of 8 categories
- 75%: participation in 3 out of 8 categories
- 100%: participation in 4 or more categories
- (main evidence) written and/or verbal participation during class
- Public work
- (main evidence) Taking notes on your GitHub class notes repositories
- (optional) Deploy an iSEE
shinyapp on shinyapps.io - (advanced and optional) R package with notes following
usethis::create_package()+biocthis::use_bioc_pkg_templates().
- Final project
- (main evidence) code in a public GitHub repository (different from the notes one) following the structure from LieberInstitute/template_project
- (optional) a rendered Rmd file (RPubs, GitHub, etc) on a public URL
- (optional advanced) Copy this repository, remove all the Rmds except the index.Rmd and edit accordingly. GitHub Actions will then help you render it. You’ll need to fetch the
gh-pagesbranch and at least make one commit for the HTML to be available such as this commit. You might also run into this GitHub default permission setting that you’ll need to change.
The final grade will be a weighted average from the evaluation by the TA (20%), your public work (40%) and your final project (40%).
Projects, public work and notes are due at 9 am US Eastern on Monday February 23rd, though we recommend doing any work related to this course by Friday February 20th.
R session information
Details on the R version used for making this book. The source code is available at lcolladotor/rnaseq_LCG-UNAM_2026.
## Load the package at the top of your script
library("sessioninfo")
## Utilities
library("BiocStyle")
library("biocthis")
library("here")
library("lobstr")
library("postcards")
library("usethis")
library("sessioninfo")
## Main containers / vis
library("SummarizedExperiment")
library("iSEE")
## RNA-seq
library("edgeR")
library("ExploreModelMatrix")
library("limma")
library("recount3")
## Visualization
library("ComplexHeatmap")
library("ggplot2")
library("patchwork")
library("pheatmap")
library("RColorBrewer")
## Advanced
library("spatialLIBD")## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.5.2 (2025-10-31)
## os Ubuntu 24.04.3 LTS
## system x86_64, linux-gnu
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz UTC
## date 2026-02-13
## pandoc 3.8.2.1 @ /usr/bin/ (via rmarkdown)
## quarto 1.7.32 @ /usr/local/bin/quarto
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## abind 1.4-8 2024-09-12 [1] RSPM (R 4.5.0)
## AnnotationDbi 1.72.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## AnnotationHub 4.0.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## attempt 0.3.1 2020-05-03 [1] RSPM (R 4.5.0)
## beachmat 2.26.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## beeswarm 0.4.0 2021-06-01 [1] RSPM (R 4.5.0)
## benchmarkme 1.0.8 2022-06-12 [1] RSPM (R 4.5.0)
## benchmarkmeData 2.0.0 2026-01-19 [1] RSPM (R 4.5.0)
## Biobase * 2.70.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## BiocFileCache 3.0.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## BiocGenerics * 0.56.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## BiocIO 1.20.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## BiocManager 1.30.27 2025-11-14 [2] CRAN (R 4.5.2)
## BiocNeighbors 2.4.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## BiocParallel 1.44.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## BiocSingular 1.26.1 2025-11-17 [1] Bioconductor 3.22 (R 4.5.2)
## BiocStyle * 2.38.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## biocthis * 1.20.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## BiocVersion 3.22.0 2025-10-07 [2] Bioconductor 3.22 (R 4.5.2)
## Biostrings 2.78.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## bit 4.6.0 2025-03-06 [1] RSPM (R 4.5.0)
## bit64 4.6.0-1 2025-01-16 [1] RSPM (R 4.5.0)
## bitops 1.0-9 2024-10-03 [1] RSPM (R 4.5.0)
## blob 1.3.0 2026-01-14 [1] RSPM (R 4.5.0)
## bookdown 0.46 2025-12-05 [1] RSPM (R 4.5.0)
## bslib 0.10.0 2026-01-26 [2] RSPM (R 4.5.0)
## cachem 1.1.0 2024-05-16 [2] RSPM (R 4.5.0)
## cigarillo 1.0.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## circlize 0.4.17 2025-12-08 [1] RSPM (R 4.5.0)
## cli 3.6.5 2025-04-23 [2] RSPM (R 4.5.0)
## clue 0.3-66 2024-11-13 [1] RSPM (R 4.5.0)
## cluster 2.1.8.2 2026-02-05 [3] RSPM (R 4.5.0)
## codetools 0.2-20 2024-03-31 [3] CRAN (R 4.5.2)
## colorspace 2.1-2 2025-09-22 [1] RSPM (R 4.5.0)
## colourpicker 1.3.0 2023-08-21 [1] RSPM (R 4.5.0)
## ComplexHeatmap * 2.26.1 2026-02-03 [1] Bioconductor 3.22 (R 4.5.2)
## config 0.3.2 2023-08-30 [1] RSPM (R 4.5.0)
## cowplot 1.2.0 2025-07-07 [1] RSPM (R 4.5.0)
## crayon 1.5.3 2024-06-20 [2] RSPM (R 4.5.0)
## curl 7.0.0 2025-08-19 [2] RSPM (R 4.5.0)
## data.table 1.18.2.1 2026-01-27 [1] RSPM (R 4.5.0)
## DBI 1.2.3 2024-06-02 [1] RSPM (R 4.5.0)
## dbplyr 2.5.1 2025-09-10 [1] RSPM (R 4.5.0)
## DelayedArray 0.36.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## digest 0.6.39 2025-11-19 [2] RSPM (R 4.5.0)
## doParallel 1.0.17 2022-02-07 [1] RSPM (R 4.5.0)
## dplyr 1.2.0 2026-02-03 [1] RSPM (R 4.5.0)
## DT 0.34.0 2025-09-02 [1] RSPM (R 4.5.0)
## edgeR * 4.8.2 2025-12-25 [1] Bioconductor 3.22 (R 4.5.2)
## evaluate 1.0.5 2025-08-27 [2] RSPM (R 4.5.0)
## ExperimentHub 3.0.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## ExploreModelMatrix * 1.22.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## farver 2.1.2 2024-05-13 [1] RSPM (R 4.5.0)
## fastmap 1.2.0 2024-05-15 [2] RSPM (R 4.5.0)
## filelock 1.0.3 2023-12-11 [1] RSPM (R 4.5.0)
## foreach 1.5.2 2022-02-02 [1] RSPM (R 4.5.0)
## fs 1.6.6 2025-04-12 [2] RSPM (R 4.5.0)
## generics * 0.1.4 2025-05-09 [1] RSPM (R 4.5.0)
## GenomicAlignments 1.46.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## GenomicRanges * 1.62.1 2025-12-08 [1] Bioconductor 3.22 (R 4.5.2)
## GetoptLong 1.1.0 2025-11-28 [1] RSPM (R 4.5.0)
## ggbeeswarm 0.7.3 2025-11-29 [1] RSPM (R 4.5.0)
## ggplot2 * 4.0.2 2026-02-03 [1] RSPM (R 4.5.0)
## ggrepel 0.9.6 2024-09-07 [1] RSPM (R 4.5.0)
## GlobalOptions 0.1.3 2025-11-28 [1] RSPM (R 4.5.0)
## glue 1.8.0 2024-09-30 [2] RSPM (R 4.5.0)
## golem 0.5.1 2024-08-27 [1] RSPM (R 4.5.0)
## gridExtra 2.3 2017-09-09 [1] RSPM (R 4.5.0)
## gtable 0.3.6 2024-10-25 [1] RSPM (R 4.5.0)
## here * 1.0.2 2025-09-15 [1] RSPM (R 4.5.0)
## htmltools 0.5.9 2025-12-04 [2] RSPM (R 4.5.0)
## htmlwidgets 1.6.4 2023-12-06 [2] RSPM (R 4.5.0)
## httpuv 1.6.16 2025-04-16 [2] RSPM (R 4.5.0)
## httr 1.4.7 2023-08-15 [1] RSPM (R 4.5.0)
## httr2 1.2.2 2025-12-08 [2] RSPM (R 4.5.0)
## igraph 2.2.2 2026-02-12 [1] RSPM (R 4.5.0)
## IRanges * 2.44.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## irlba 2.3.7 2026-01-30 [1] RSPM (R 4.5.0)
## iSEE * 2.22.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## iterators 1.0.14 2022-02-05 [1] RSPM (R 4.5.0)
## jquerylib 0.1.4 2021-04-26 [2] RSPM (R 4.5.0)
## jsonlite 2.0.0 2025-03-27 [2] RSPM (R 4.5.0)
## KEGGREST 1.50.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## knitr 1.51 2025-12-20 [2] RSPM (R 4.5.0)
## later 1.4.5 2026-01-08 [2] RSPM (R 4.5.0)
## lattice 0.22-9 2026-02-09 [3] RSPM (R 4.5.0)
## lazyeval 0.2.2 2019-03-15 [1] RSPM (R 4.5.0)
## lifecycle 1.0.5 2026-01-08 [1] RSPM (R 4.5.0)
## limma * 3.66.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## listviewer 4.0.0 2023-09-30 [1] RSPM (R 4.5.0)
## lobstr * 1.1.3 2025-11-14 [1] RSPM (R 4.5.0)
## locfit 1.5-9.12 2025-03-05 [1] RSPM (R 4.5.0)
## magick 2.9.0 2025-09-08 [1] RSPM (R 4.5.0)
## magrittr 2.0.4 2025-09-12 [2] RSPM (R 4.5.0)
## MASS 7.3-65 2025-02-28 [3] CRAN (R 4.5.2)
## Matrix 1.7-4 2025-08-28 [3] CRAN (R 4.5.2)
## MatrixGenerics * 1.22.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## matrixStats * 1.5.0 2025-01-07 [1] RSPM (R 4.5.0)
## memoise 2.0.1 2021-11-26 [2] RSPM (R 4.5.0)
## mgcv 1.9-4 2025-11-07 [3] RSPM (R 4.5.0)
## mime 0.13 2025-03-17 [2] RSPM (R 4.5.0)
## miniUI 0.1.2 2025-04-17 [2] RSPM (R 4.5.0)
## nlme 3.1-168 2025-03-31 [3] CRAN (R 4.5.2)
## otel 0.2.0 2025-08-29 [2] RSPM (R 4.5.0)
## paletteer 1.7.0 2026-01-08 [1] RSPM (R 4.5.0)
## patchwork * 1.3.2 2025-08-25 [1] RSPM (R 4.5.0)
## pheatmap * 1.0.13 2025-06-05 [1] RSPM (R 4.5.0)
## pillar 1.11.1 2025-09-17 [2] RSPM (R 4.5.0)
## pkgconfig 2.0.3 2019-09-22 [2] RSPM (R 4.5.0)
## plotly 4.12.0 2026-01-24 [1] RSPM (R 4.5.0)
## png 0.1-8 2022-11-29 [1] RSPM (R 4.5.0)
## postcards * 0.2.3 2022-01-07 [1] RSPM (R 4.5.0)
## promises 1.5.0 2025-11-01 [2] RSPM (R 4.5.0)
## purrr 1.2.1 2026-01-09 [2] RSPM (R 4.5.0)
## R.cache 0.17.0 2025-05-02 [1] RSPM (R 4.5.0)
## R.methodsS3 1.8.2 2022-06-13 [1] RSPM (R 4.5.0)
## R.oo 1.27.1 2025-05-02 [1] RSPM (R 4.5.0)
## R.utils 2.13.0 2025-02-24 [1] RSPM (R 4.5.0)
## R6 2.6.1 2025-02-15 [2] RSPM (R 4.5.0)
## rappdirs 0.3.4 2026-01-17 [2] RSPM (R 4.5.0)
## RColorBrewer * 1.1-3 2022-04-03 [1] RSPM (R 4.5.0)
## Rcpp 1.1.1 2026-01-10 [2] RSPM (R 4.5.0)
## RCurl 1.98-1.17 2025-03-22 [1] RSPM (R 4.5.0)
## recount3 * 1.20.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## rematch2 2.1.2 2020-05-01 [1] RSPM (R 4.5.0)
## restfulr 0.0.16 2025-06-27 [1] RSPM (R 4.5.2)
## rintrojs 0.3.4 2024-01-11 [1] RSPM (R 4.5.0)
## rjson 0.2.23 2024-09-16 [1] RSPM (R 4.5.0)
## rlang 1.1.7 2026-01-09 [1] RSPM (R 4.5.0)
## rmarkdown 2.30 2025-09-28 [2] RSPM (R 4.5.0)
## rprojroot 2.1.1 2025-08-26 [2] RSPM (R 4.5.0)
## Rsamtools 2.26.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## RSQLite 2.4.6 2026-02-06 [1] RSPM (R 4.5.0)
## rstudioapi 0.18.0 2026-01-16 [2] RSPM (R 4.5.0)
## rsvd 1.0.5 2021-04-16 [1] RSPM (R 4.5.0)
## rtracklayer 1.70.1 2025-12-22 [1] Bioconductor 3.22 (R 4.5.2)
## S4Arrays 1.10.1 2025-12-01 [1] Bioconductor 3.22 (R 4.5.2)
## S4Vectors * 0.48.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## S7 0.2.1 2025-11-14 [1] RSPM (R 4.5.0)
## sass 0.4.10 2025-04-11 [2] RSPM (R 4.5.0)
## ScaledMatrix 1.18.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## scales 1.4.0 2025-04-24 [1] RSPM (R 4.5.0)
## scater 1.38.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## scuttle 1.20.0 2025-10-30 [1] Bioconductor 3.22 (R 4.5.2)
## Seqinfo * 1.0.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## sessioninfo * 1.2.3 2025-02-05 [2] RSPM (R 4.5.0)
## shape 1.4.6.1 2024-02-23 [1] RSPM (R 4.5.0)
## shiny 1.12.1 2025-12-09 [2] RSPM (R 4.5.0)
## shinyAce 0.4.4 2025-02-03 [1] RSPM (R 4.5.0)
## shinydashboard 0.7.3 2025-04-21 [1] RSPM (R 4.5.0)
## shinyjs 2.1.1 2026-01-15 [1] RSPM (R 4.5.0)
## shinyWidgets 0.9.0 2025-02-21 [1] RSPM (R 4.5.0)
## SingleCellExperiment * 1.32.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## SparseArray 1.10.8 2025-12-18 [1] Bioconductor 3.22 (R 4.5.2)
## SpatialExperiment * 1.20.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## spatialLIBD * 1.22.0 2025-11-04 [1] Bioconductor 3.22 (R 4.5.2)
## statmod 1.5.1 2025-10-09 [1] RSPM (R 4.5.0)
## styler 1.11.0 2025-10-13 [1] RSPM (R 4.5.0)
## SummarizedExperiment * 1.40.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## tibble 3.3.1 2026-01-11 [2] RSPM (R 4.5.0)
## tidyr 1.3.2 2025-12-19 [1] RSPM (R 4.5.0)
## tidyselect 1.2.1 2024-03-11 [1] RSPM (R 4.5.0)
## usethis * 3.2.1 2025-09-06 [2] RSPM (R 4.5.0)
## vctrs 0.7.1 2026-01-23 [1] RSPM (R 4.5.0)
## vipor 0.4.7 2023-12-18 [1] RSPM (R 4.5.0)
## viridis 0.6.5 2024-01-29 [1] RSPM (R 4.5.0)
## viridisLite 0.4.3 2026-02-04 [1] RSPM (R 4.5.0)
## withr 3.0.2 2024-10-28 [2] RSPM (R 4.5.0)
## xfun 0.56 2026-01-18 [2] RSPM (R 4.5.0)
## XML 3.99-0.22 2026-02-10 [1] RSPM (R 4.5.0)
## xtable 1.8-4 2019-04-21 [2] RSPM (R 4.5.0)
## XVector 0.50.0 2025-10-29 [1] Bioconductor 3.22 (R 4.5.2)
## yaml 2.3.12 2025-12-10 [2] RSPM (R 4.5.0)
##
## [1] /__w/_temp/Library
## [2] /usr/local/lib/R/site-library
## [3] /usr/local/lib/R/library
## * ── Packages attached to the search path.
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────## user system elapsed
## 11.458 0.927 12.014This book was last updated at 2026-02-13 16:13:18.318598.
License

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.