2 Data Science guidance sessions
2.1 Quick DSgs sign up
You can think about the Data Science guidance sessions (DSgs) as open office hours. Also for several of us (Nick, Louise, and Leo), these DSgs’es are part of our job at LIBD. So please don’t think that you are imposing in any way: we want to help you! Here are the quick links for signing up:

Leo’s team:
- Manisha Barse
- Sign up via Calendly
- Nicholas J. Eagles
- Sign up via Calendly
- Louise A. Huuki-Myers
- Sign up via Calendly
- Leonardo Collado-Torres
- Sign up via Calendly
More DSgs guides:
Further below you’ll see these links again after we:
- explain the background behind DSgs,
- some rules we have,
- how you can help us help you,
- how to frame a question,
- information we might need,
- which are our research interests and how you can sign up.
Finally, if you want to be a DSgs guide and be listed on this page, we have some information for you.
2.2 Background
Data is abundant in many aspects of our research work thanks to high-throughput experimental assays. While some analyses will require a significant amount of time from people who primarily do that type of work, we know that many of us are interested in learning more about working with data. As such, as a team, the DSgs-guides will provide Data Science guidance sessions (DSgs).
These are 25 minute sessions where we’ll try our best to help you, teach you, troubleshoot with you, and overall guide you as much as we can. You can think of DSgs as open office hours. It is how we contribute to the LIBD’s data science capacity growing goals, and for several of us, providing DSgs sessions is part of our job at LIBD.
As with any office hours in a class, DSgs sessions are particularly helpful if you want to work on a data problem or gain some data skills across a period of weeks or months. They do not work well for last minute requests. Working with data and programming languages can have a steep learning curve and it can feel daunting. As a team, we have trained ourselves to have a common baseline and can help you get familiarized with the concept of a project-oriented workflow. This involves tools such as RStudio, git/GitHub, how to organize your code, how to ask for help, and overall tools to make your work more reproducible. These ideas when put into practice are very helpful for finding why things are not working, as we all inevitably encounter errors in our code and the open source software we rely on. Sometimes we won’t know the answer either, but maybe we can help you construct more specific Google search terms.
Check the following slides for some of the initial ideas behind these sessions.
Our philosophy and overview on the help we provide was described in this video and slides.
2.3 Interaction rules
In order for DSgs sessions to run smoothly, it’s important that we have clear expectations of what we’ll do. DSgs sessions are not consulting sessions. We will help you as much as we can during the session, but after it’s over, we won’t spend time writing new code for you. That is, we will not take requests for work you need help finishing regardless of how important it is. That’s why we recommend planning ahead and even having weekly DSgs sessions if you have a deadline in mind instead of approaching us for help the week the project is due. We are most helpful early in a project cycle than at the end.
We understand that you might be stressed or under significant pressure, but please follow our code of conduct 3. Otherwise, we will forced to stop meeting with you.
2.4 Help us help you
When you sign up for a session, you will fill a small form where you can provide some information about why you want to meet. We will spend up to 15 minutes preparing for a session, and in order to make better use of that time, it is important that you provide us enough information so we can do a better job helping you.
2.4.1 Framing a question
Just a reminder that you should read 3 💎s by @rdpeng
— 🇲🇽 Leonardo Collado-Torres (@lcolladotor) June 2, 2022
* What is the question? https://t.co/Aet2wvBP5c w/ @jtleek
* Tukey, Design Thinking, and Better Questions https://t.co/xyh5QZtnpQ s/o @hspter
* Types of questions https://t.co/gqyJyTK8Zd w/ @elizabethmatsui #DataScience ❓ pic.twitter.com/tXlioGa3n9
Instead of saying “I need help”, it will be useful for us if you can be more specific. For example, “I need help with jaffelab::cleaningY()” or “I need help installing the Bioconductor package SummarizedExperiment on a Windows machine”. The first example lets us know that exact function you need help with, so we can revise the documentation before hand and come prepared to the meeting. The second example mentions a specific R package, where to find it (Bioconductor), and what operating system you are using. Here are some more example help requests:
- I want to learn how to use R, where should I start?
- I finished the CBDS lesson on using GitHub but I need help using git at JHPCE.
- I have some data that I generated and want to explore it. Which plots should I make?
- A collaborator suggested a specific linear regression model (
~ age * brain region), can you explain to me how to interpret the results? - Which R package should I use for performing a gene ontology enrichment analysis?
- I’m interesting in using software
xxyou wrote, can you help me understand how to doyywith it? - I like the visualization you made for project
xx, can you help me make a similar one with my data? - I want to adapt code
xxyour team wrote in the past and use it in my project, can you help me get started
Note that we’ll use the information you provided us for our DSgs session reports. We will keep your name anonymous and remove any private information.
2.4.2 Useful information
As problems get more specific, we will require more information. If you are encountering an R error that you need help with, providing a small reproducible example will make the whole experience much smoother. The R package that is very useful for providing such examples is reprex. You can watch our LIBD rstats club video on reprex to get more familiarized with this package.
To share code with us,
- Please create a
gistwhich can be either public or private. In thegistinclude all the commands you ran and the output you got. Or provide thereprexcode (preferred option). - If you are already using GitHub for version controlling your code, then include the link to the script you are running.
- If you have a companion log file at JHPCE, please share the full path to the log file 4
- Ideally include the R session information which you can generate using
options(width = 120); sessioninfo::session_info(). This information will be particularly useful for solving issues with cutting edge R/Bioconductor/GitHub packages.
2.5 Sign up
If you are interested in a 25 minute Data Science guidance session, please sign up through the Calendly links below with a session with one of our guides. If this is the first time you are using this or might need more general guidance, sign up with Leo.
If you have a Zoom account, please add the Zoom details to the calendar invitation 5.
Finally, please sign up for a maximum of two sessions per week. This will help us keep the DSgs system fair for everyone to use. Thank you!
Please try reaching out to multiple people instead of one person for all your questions. Thank you!
2.5.1 Leo’s team
- Manisha Barse (BioC Support)
- Research interests: bulk RNA-seq, scRNA-seq, building computational pipelines.
- Technologies I use: linux, python, R, github.
- Learning about: spatial transcriptomics, coding workflows.
- Sign up via Calendly
- Nicholas J. Eagles (BioC Support)
- Research interests: spatial transcriptomics, building computational pipelines, data visualization.
- Technologies I use: R/Bioconductor, Git, Nextflow, Python.
- Learning about: deep learning, LLMs for data science.
- Sign up via Calendly
- Louise A. Huuki-Myers (BioC Support)
- Research interests: bulk and single cell RNA-seq, spatial transcriptomics, RNA scope data, DEGs, data visualization.
- Technologies I use: R (tidyverse, ggplot, dplyr), Python, GitHub, Synapse upload.
- Learning about: Bioconductor Package development.
- Sign up via Calendly
- Leonardo Collado-Torres (BioC Support)
- Research interests: RNA-seq, spatial transcriptomics, un-annotated transcriptome, deconvolution, multi-omics.
- Technologies I use: R/Bioconductor, GitHub, SLURM/JHPCE.
- Learning about: team management, grant applications.
- Sign up via Calendly
2.5.2 More DSgs guides
- Madhavi Tippani (BioC Support)
- Research interests: spatial transcriptomics, calcium imaging, RNAscope, medical image processing.
- Technologies I use: Matlab, R, GitHub, SLURM/JHPCE.
- Learning about: Machine Learning, spatial transcriptomics, statistical analysis.
- Sign up via Calendly
2.6 DSgs session reports
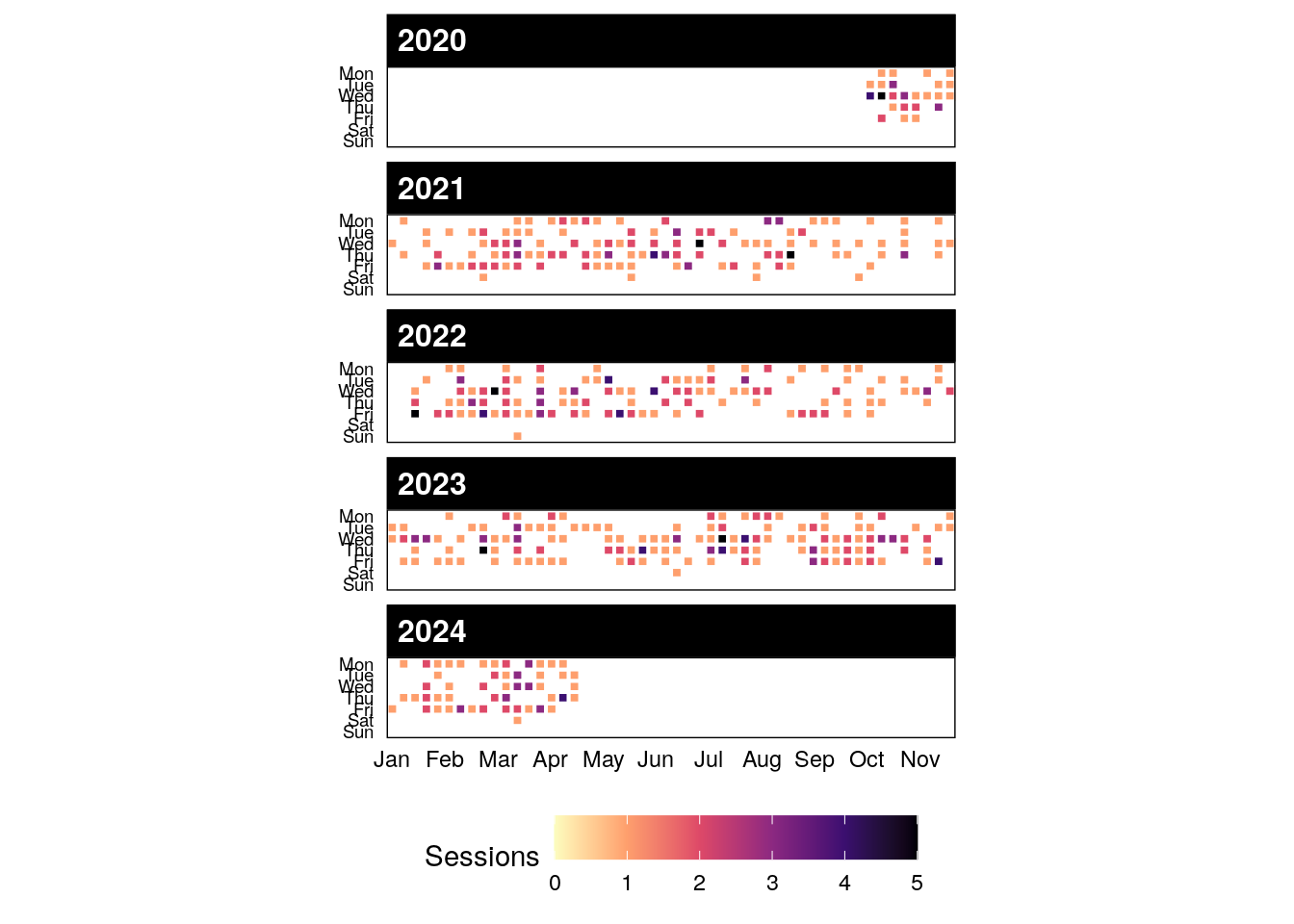
If you are interested in public information from our past sessions, check DSgs_logs, and in particular, some of our graphs.
2.7 Become a DSgs guide
Non-team members are welcome to join the DSgs-guides. This involves a significant time commitment (~ 7 hours per week) and you need to discuss this with your supervisor. These commitments are:
- helping others through DSgs sessions ~ 200 minutes per week 6
- a weekly 30 minute meeting as explained in DSgs: feedback
- attending the LIBD rstats club (1 hour per week)
- time for self-learning and/or building common resources (~ 1 hour per week)
- (recommended) practice and learn how to help others through the Bioconductor Support: practice grounds (~ 30 min per week)
Additionally, we might have some additional training sessions.
Some potential advantages are:
- access to all our training material, explained in detail at DSgs-guides
- a common structure for helping others at LIBD/JHU
- recognition through our DSgs session reports
We will update the code of conduct once Bioconductor publishes their latest code of conduct in the Fall of 2020.↩︎
You might want to use the slurmjobs package for helping you create such log files.↩︎
If you are using Google Calendar, there is a Google Calendar Zoom integration. The Outlook version LIBD uses does not support the Zoom integration, so you’ll have to add the information manually to the calendar invitation.↩︎
We recommend four 25 minute sessions, each with 15 minutes of preparation and 10 minutes of post-meeting wrap up.↩︎