2 smokingMouse study
Instructor: Daianna
As part of your training, we’ll guide you through an analysis with real bulk RNA-seq data.
The dataset that we’ll be using is a RangedSummarizedExperiment coming from the smokingMouse (Gonzalez-Padilla and Collado-Torres, 2023) package, but in this chapter we’ll explain you the study design and the experimental stages that preceded the data generation.
The smoking mouse study is a project currently being developed at the Lieber Institute for Brain Development by Daianna Gonzalez-Padilla and Leonardo Collado-Torres.
2.1 Introduction
Nowadays, maternal smoking during pregnancy (MSDP) is one of the major health concerns worldwide not only because of the effects on the smoker but because the health of the offspring could be dramatically affected by smoking exposure, particularly their cognitive and behavioral traits. But are those effects caused by cigarette smoke only? Or are those alterations given by a specific component of cigarettes such as nicotine? The latter is a very relevant question because most of the studies have focused on analyzing smoking effects, ignoring the role of nicotine, an active component of the cigarettes. Understanding which effects are specifically dictated by nicotine prenatal exposure will enable more directed studies of this drug on the developing brain, once affected genes have been identified and experimentally proven.
2.2 Overview
This study consisted of a series of bulk RNA-sequencing experiments performed on brain samples from adult mice and pups and on blood samples from adults. Adult mice were either exposed to cigarette smoke (what we call smoking experiment and smoking mice) or controls of the smoking experiment, or they were administered nicotine (nicotine experiment and nicotine mice) or controls of the nicotine experiment, and they were either pregnant or not. Smoking, nicotine and control pups are pups that were born to smoking, nicotine and control pregnant mice, respectively. This experimental design allowed us to contrast the altered features in both conditions and to compare the effects in different mouse ages, sexes, tissues and pregnancy states.
The original datasets contain gene, transcript, exon, and exon-exon junction expression levels across 208 samples, but in this course we’ll only use gene data in nicotine samples to simplify the analyses.
2.3 Goals
This project actually aimed to answer 4 questions, all related to smoking and nicotine effects in mouse.
2.3.1 Main objective
The main goal was to identify and differentiate the effects of prenatal nicotine and smoking exposures on gene, transcript, exon and junction expression of developing brain of pups. Basically, to perform a differential expression analysis on mouse data to find significant feature expression changes.
2.3.2 Secondary objectives
- To evaluate the affected genes by each substance on adult brain in order to compare pup and adult results.
- To examine the effects of smoking on adult blood and brain to search for overlapping biomarkers in both tissues (Can blood analyses capture environmental effects on brain?)
- To compare mouse differentially expressed genes (DEGs) with results from previous studies in human (Semick, S.A. et al. (2018) and Toikumo, S. et al. (2023)).
Check here for the code of the original and complete analyses done.
2.4 Study design
As presented in Figure 1: A), 36 pregnant dams and 35 non-pregnant female adult mice were either administered nicotine by intraperitoneal injection (IP; n=12), exposed to cigarette smoke in smoking chambers (n=24), or controls (n=35; 11 nicotine controls and 24 smoking controls). A total of 137 pups were born to pregnant dams: 19 were born to mice that were administered nicotine, 46 to mice exposed to cigarette smoke and the remaining 72 to control mice (23 to nicotine controls and 49 to smoking controls). Samples from frontal cortices of P0 pups and adults were obtained, as well as blood samples from smoking-exposed and smoking control adults. Then, as shown in B) RNA was extracted from all those samples and RNA-seq libraries were prepared and sequenced to obtain expression counts for genes, exons, transcripts, and exon-exon junctions.
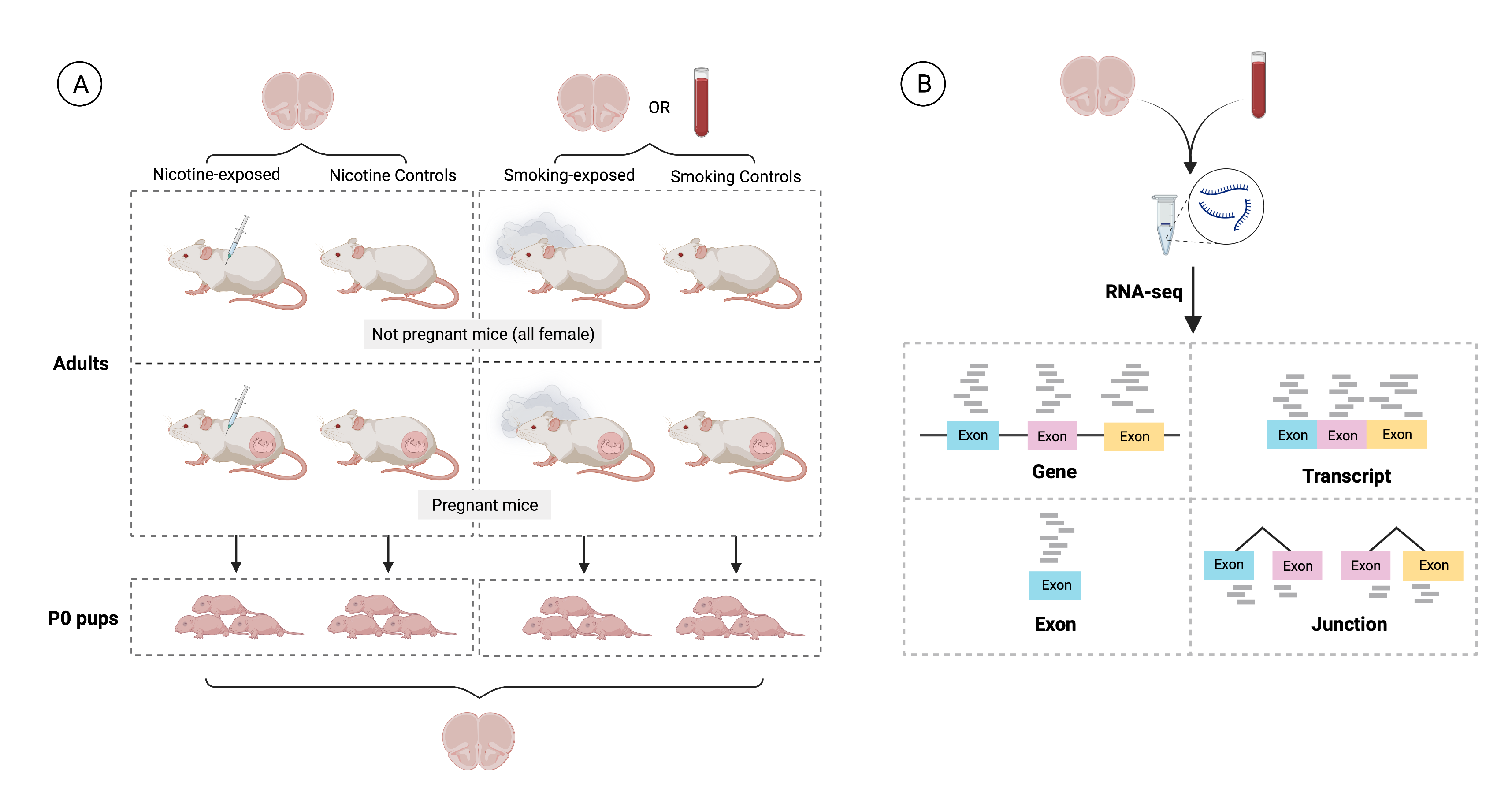
Figure 1: Experimental design of the study.