Publications
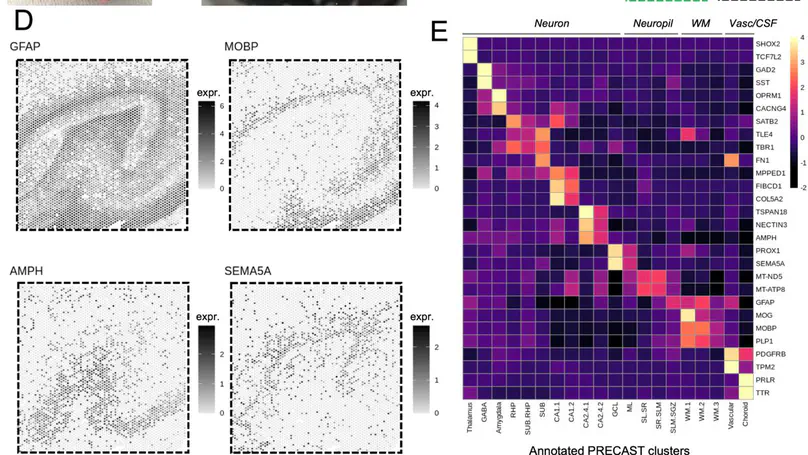
Cell types in the hippocampus with unique morphology, physiology and connectivity serve specialized functions associated with cognition and mood. These cell types are spatially organized, necessitating molecular profiling strategies that retain cytoarchitectural organization. Here we generated spatially-resolved transcriptomics (SRT) and single-nucleus RNA-sequencing (snRNA-seq) data from anterior human hippocampus in ten adult neurotypical donors. Using non-negative matrix factorization (NMF) and label transfer, we integrated these data by defining gene expression patterns within the snRNA-seq data and then inferring expression in the SRT data. These patterns captured transcriptional variation across neuronal cell types and indicated spatial organization of excitatory and inhibitory postsynaptic specializations. Leveraging the NMF and label transfer approach with rodent datasets, we identified putative patterns of activity-dependent transcription and circuit connectivity in the human SRT dataset. Finally, we characterized the spatial organization of NMF patterns corresponding to pyramidal neurons and identified regionally-specific snRNA-seq clusters of the retrohippocampus, subiculum and presubiculum. To make this molecular atlas widely accessible, raw and processed data are freely available, including through interactive web applications.
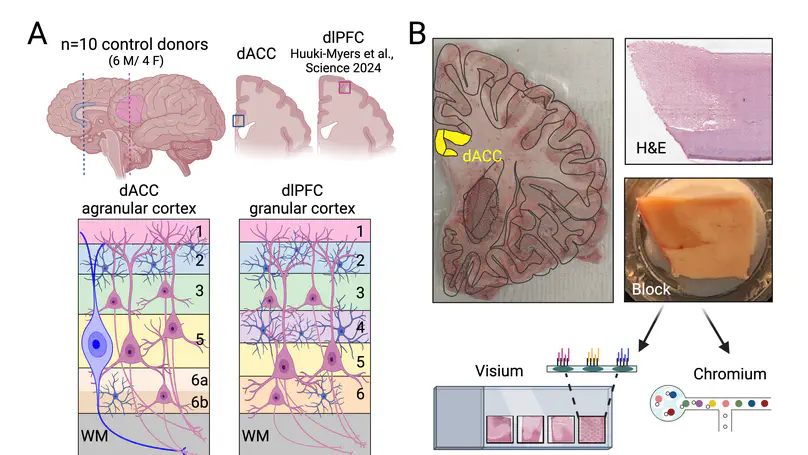
In the human brain, the dorsal anterior cingulate cortex (dACC) plays key roles in various components of cognitive control, and is particularly relevant for reward processing and conflict monitoring. The dACC regulates expression of fear and pain, and its dysfunction is implicated in a number of neuropsychiatric disorders. Compared to more recently specialized neocortical areas, such as the dorsolateral prefrontal cortex (dlPFC), the dACC is evolutionarily older. The region’s agranular structure, and other evolutionary specializations, such as the presence of von Economo neurons (VENs), contribute to its specialized roles in cognitive and emotional processing. Here, we generated paired spatially-resolved transcriptomics (SRT) and single-nucleus RNA-sequencing (snRNA-seq) data from adjacent tissue sections of the dACC in ten adult neurotypical donors to define molecular profiles for dACC cell types and spatial domains. Using non-negative matrix factorization (NMF), we integrated these data by identifying gene expression patterns within the snRNA-seq data, which were projected onto the SRT data to infer the spatial localization. Combining these data with publicly available resources, we revealed insights about molecular profiles, spatial topography, enrichment of disease risk, and putative connectivity of spatially-localized dACC cell types, including VENs. Utilizing published dlPFC snRNA-seq and SRT data collected in the same neurotypical brain donors used here, we deployed cross-region comparison analyses between dACC and dlPFC to understand spatio-molecular specializations and laminar organization across human brain evolution. To make this comprehensive molecular resource accessible to the scientific community, we made both raw and processed data freely available, including through interactive web applications.
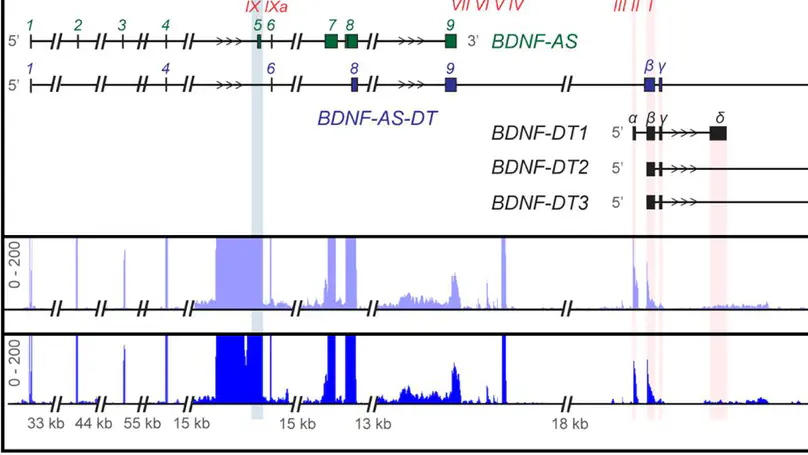
Divergent transcription from bidirectional promoters is frequently observed in eukaryotic genomes, but the biological relevance of divergent RNA transcripts (DT) is unknown. We identified and characterized BDNF-DT, a novel DT gene, and BDNF-AS-DT, a novel readthrough gene, in the locus containing BDNF, a gene with key roles in neuronal development, differentiation, and synaptic plasticity. BDNF-DT is independent from the known BDNF antisense (BDNF-AS), and its expression is developmentally regulated and positively correlated with BDNF in human postmortem dorsolateral prefrontal cortex (DLPFC). BDNF-DT and BDNF-AS-DT expression increase after induced depolarization, but the temporal dynamics follow expression of BDNF, suggesting a regulatory role. Moreover, CRISPR-mediated upregulation of BDNF in human neural progenitor cells drives BDNF-DT expression. Finally, BDNF-DT shows higher expression in DLPFC from patients diagnosed with schizophrenia compared to neurotypical controls, and genetically predicted lower expression of the BDNF-AS-DT readthrough transcript is associated with schizophrenia and with the schizophrenia-associated C allele of the rs6265 single-nucleotide polymorphism. These data suggest that BDNF-DT and BDNF-AS-DT contribute to BDNF regulation and schizophrenia risk.
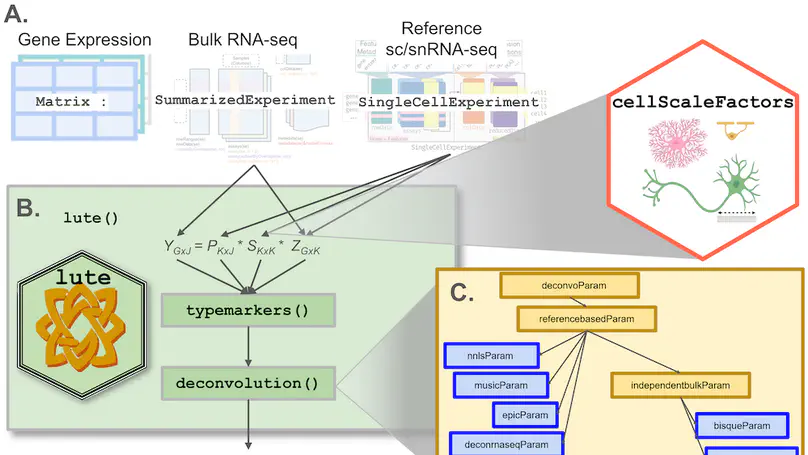
Background: Relative cell type fraction estimates in bulk RNA-sequencing data are important to control for cell composition differences across heterogenous tissue samples. While there exist algorithms to estimate the cell type proportions in tissues, a major challenge is the algorithms can show reduced performance if using tissues that have varying cell sizes, such as in brain tissue. In this way, without adjusting for differences in cell sizes, computational algorithms estimate the relative fraction of RNA attributable to each cell type, rather than the relative fraction of cell types, leading to potentially biased estimates in cellular composition. Furthermore, these tools were built on different frameworks with non-uniform input data formats while addressing different types of systematic errors or unwanted bias. Results: We present lute, a software tool to accurately deconvolute cell types with varying sizes. Our package lute wraps existing deconvolution algorithms in a flexible and extensible framework to enable easy benchmarking and comparison of existing deconvolution algorithms. Using simulated and real datasets, we demonstrate how lute adjusts for differences in cell sizes to improve the accuracy of cell composition. Conclusions: Our software (https://bioconductor.org/packages/lute) can be used to enhance and improve existing deconvolution algorithms and can be used broadly for any type of tissue containing cell types with varying cell sizes.
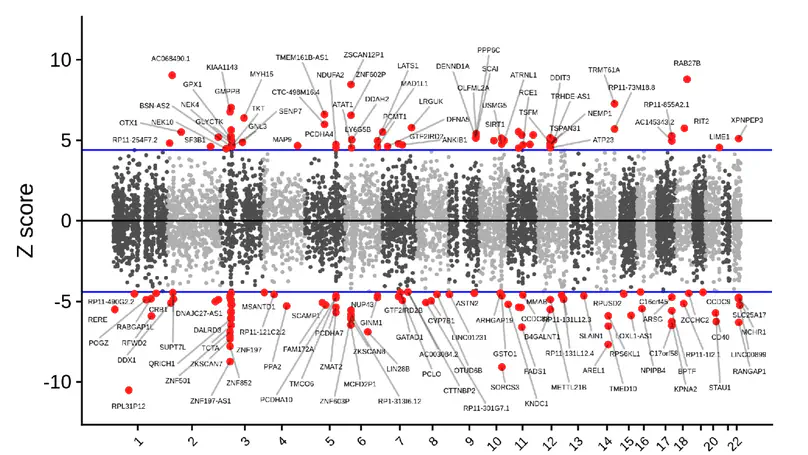
Major Depressive Disorder (MDD) is a common, complex disorder that is a leading cause of disability worldwide and a significant risk factor for suicide. In this study, we have performed the largest molecular analysis of MDD in postmortem human brains (846 samples across 458 individuals) in the subgenual Anterior Cingulate Cortex (sACC) and the Amygdala, two regions central to mood regulation and the pathophysiology of MDD. We found extensive expression differences, particularly at the level of specific transcripts, with prominent enrichment for genes associated with the vesicular functioning, the postsynaptic density, GTPase signaling, and gene splicing. We find associated transcriptional features in 107 of 243 genome-wide significant loci for MDD and, through integrative analyses, highlight convergence of genetic risk, gene expression, and network-based analyses on dysregulated glutamatergic signaling and synaptic vesicular functioning. Together, these results provide an initial mechanistic understanding of MDD and highlight potential targets for novel drug discovery.
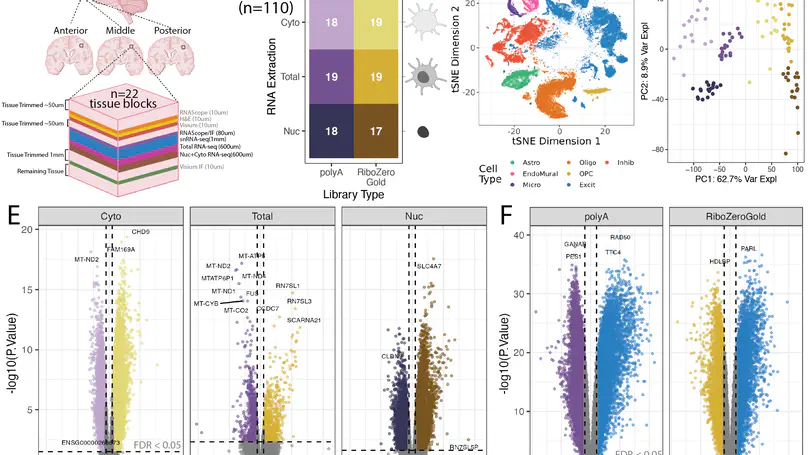
Cellular deconvolution of bulk RNA-sequencing data using single cell/nuclei RNA-seq reference data is an important strategy for estimating cell type composition in heterogeneous tissues, such as the human brain. Here, we generate a multi-assay dataset in postmortem human dorsolateral prefrontal cortex from 22 tissue blocks, including bulk RNA-seq, reference snRNA-seq, and orthogonal measurement of cell type proportions with RNAScope/ImmunoFluorescence. We use this dataset to evaluate six deconvolution algorithms. Bisque and hspe were the most accurate methods. The dataset, as well as the Mean Ratio gene marker finding method, is made available in the DeconvoBuddies R/Bioconductor package.
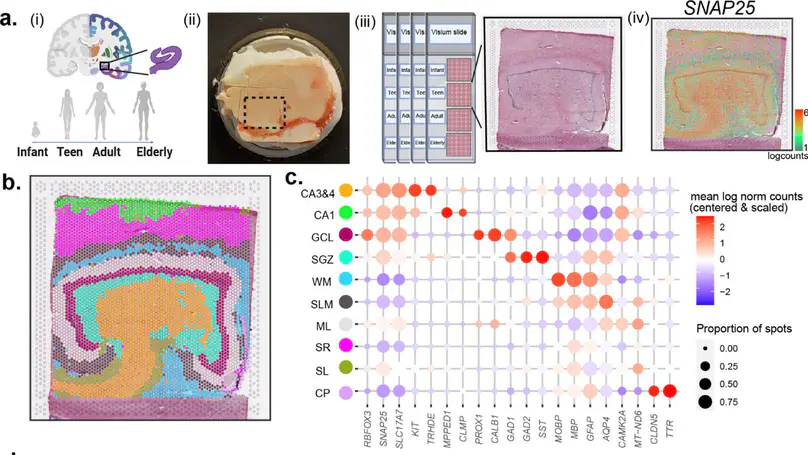
The dentate gyrus of the hippocampus is important for many cognitive functions, including learning, memory, and mood. Here, we present transcriptome-wide spatial gene expression maps of the human dentate gyrus and investigate age-associated changes across the lifespan. Genes associated with neurogenesis and the extracellular matrix are enriched in infants and decline throughout development and maturation. Following infancy, inhibitory neuron markers increase, and cellular proliferation markers decrease. We also identify spatio-molecular signatures that support existing evidence for protracted maturation of granule cells during adulthood and age-associated increases in neuroinflammation-related gene expression. Our findings support the notion that the hippocampal neurogenic niche undergoes major changes following infancy and identify molecular regulators of brain aging in glial- and neuropil-enriched tissue.
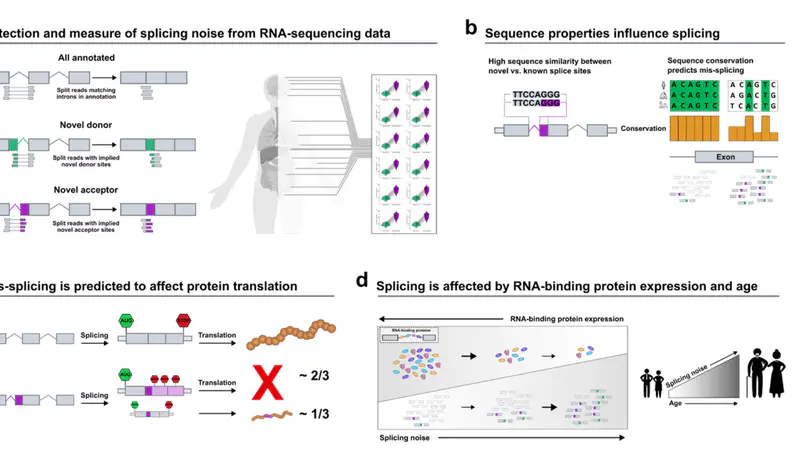
Alternative splicing impacts most multi-exonic human genes. Inaccuracies during this process may have an important role in ageing and disease. Here, we investigate splicing accuracy using RNA-sequencing data from {\textgreater}14k control samples and 40 human body sites, focusing on split reads partially mapping to known transcripts in annotation. We show that splicing inaccuracies occur at different rates across introns and tissues and are affected by the abundance of core components of the spliceosome assembly and its regulators. We find that age is positively correlated with a global decline in splicing fidelity, mostly affecting genes implicated in neurodegenerative diseases. We find support for the latter by observing a genome-wide increase in splicing inaccuracies in samples affected with Alzheimer’s disease as compared to neurologically normal individuals. In this work, we provide an in-depth characterisation of splicing accuracy, with implications for our understanding of the role of inaccuracies in ageing and neurodegenerative disorders
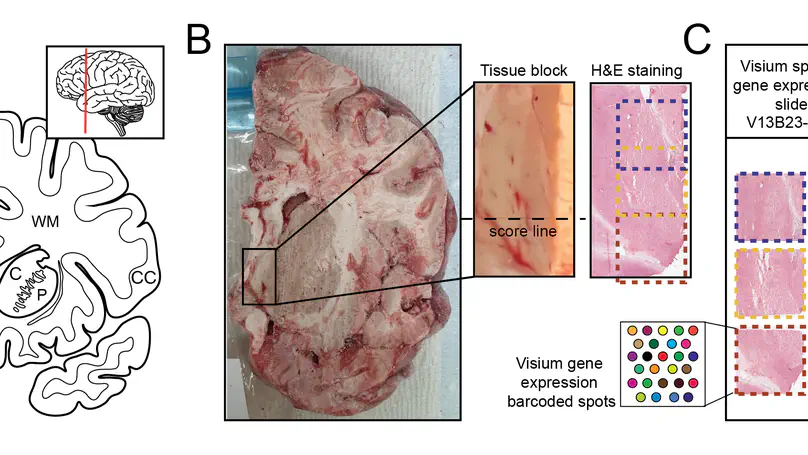
Background Visium is a widely-used spatially-resolved transcriptomics assay available from 10x Genomics. Standard Visium capture areas (6.5mm by 6.5mm) limit the survey of larger tissue structures, but combining overlapping images and associated gene expression data allow for more complex study designs. Current software can handle nested or partial image overlaps, but is designed for merging up to two capture areas, and cannot account for some technical scenarios related to capture area alignment. Results We generated Visium data from a postmortem human tissue sample such that two capture areas were partially overlapping and a third one was adjacent. We developed the R/Bioconductor package visiumStitched, which facilitates stitching the images together with Fiji (ImageJ), and constructing SpatialExperiment R objects with the stitched images and gene expression data. visiumStitched constructs an artificial hexagonal array grid which allows seamless downstream analyses such as spatially-aware clustering without discarding data from overlapping spots. Data stitched with visiumStitched can then be interactively visualized with spatialLIBD. Conclusions visiumStitched provides a simple, but flexible framework to handle various multi-capture area study design scenarios. Specifically, it resolves a data processing step without disrupting analysis workflows and without discarding data from overlapping spots. visiumStitched relies on affine transformations by Fiji, which have limitations and are less accurate when aligning against an atlas or other situations. visiumStitched provides an easy-to-use solution which expands possibilities for designing multi-capture area study designs.
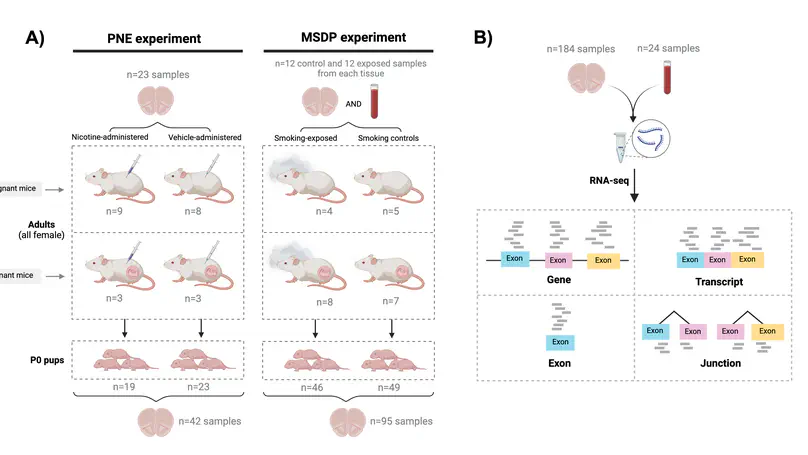
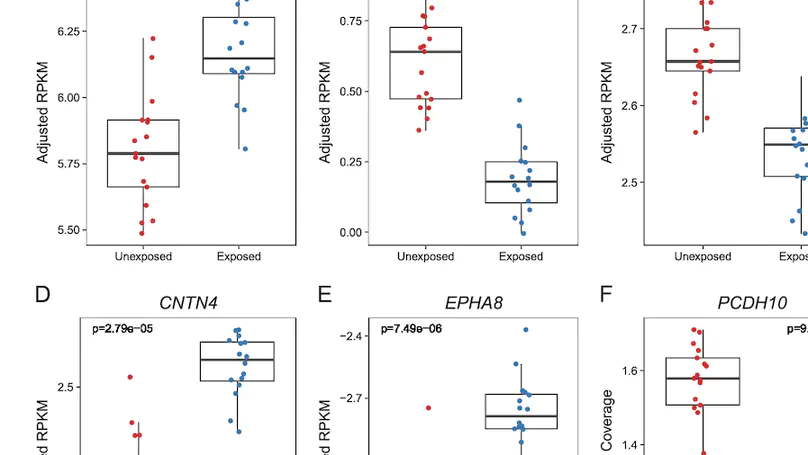
Maternal smoking during pregnancy (MSDP) is associated with significant cognitive and behavioral effects on offspring. While neurodevelopmental outcomes have been studied for prenatal exposure to nicotine, the main psychoactive component of cigarette smoke, its contribution to MSDP effects has never been explored. Comparing the effects of these substances on molecular signaling in the prenatal and adult brain may provide insights into nicotinic and broader tobacco consequences that are developmental-stage specific or age-independent. Pregnant mice were administered nicotine or exposed to chronic cigarette smoke, and RNA-sequencing was performed on frontal cortices of postnatal day 0 pups born to these mice, as well as on frontal cortices and blood of the adult dams. We identified 1,010 and 4,165 differentially expressed genes (DEGs) in nicotine and smoking-exposed pup brains, respectively (FDR<0.05, Ns = 19 nicotine-exposed vs 23 vehicle-exposed; 46 smoking-exposed vs 49 controls). Prenatal nicotine exposure (PNE) alone was related to dopaminergic synapses and long-term synaptic depression, whereas MSDP was associated with the SNARE complex and vesicle transport. Both substances affected SMN-Sm protein complexes and postsynaptic endosomes. Analyses at the transcript, exon, and exon-exon junction levels supported gene level results and revealed additional smoking-affected processes. No DEGs at FDR<0.05 were found in adult mouse brain for any substance (12 nicotine-administered vs 11 vehicle-administered; 12 smoking-exposed vs 12 controls), nor in adult blood (12 smoking-exposed vs 12 controls), and only 3% and 6.41% of the DEGs in smoking-exposed pup brain replicated in smoking-exposed blood and human prenatal brain, respectively. Together, these results demonstrate variable but overlapping molecular effects of PNE and MSDP on the developing brain, and attenuated effects of both smoking and nicotine on adult versus fetal brain.
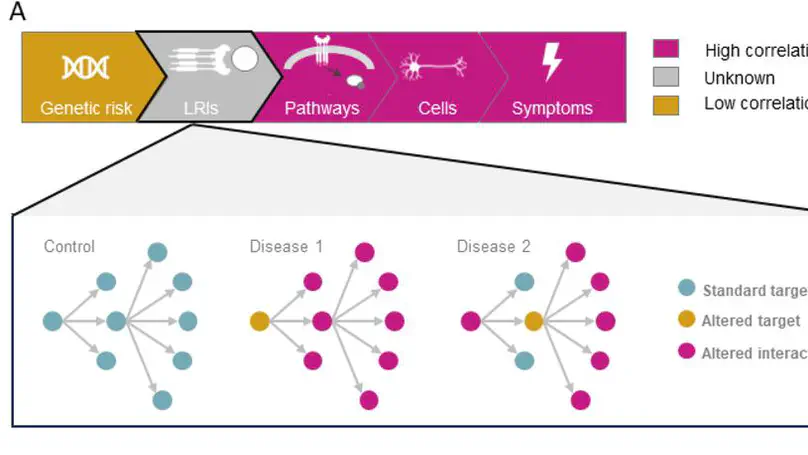
Neurodegenerative disorders have overlapping symptoms and have high comorbidity rates, but this is not reflected in overlaps of risk genes. We have investigated whether ligand-receptor interactions (LRIs) are a mechanism by which distinct genes associated with disease risk can impact overlapping outcomes. We found that LRIs are likely disrupted in neurological disease and that the ligand-receptor networks associated with neurological diseases have substantial overlaps. Specifically, 96.8% of LRIs associated with disease risk are interconnected in a single LR network. These ligands and receptors are enriched for roles in inflammatory pathways and highlight the role of glia in cross-disease risk. Disruption to this LR network due to disease-associated processes (e.g. differential transcript use, protein misfolding) is likely to contribute to disease progression and risk of comorbidity. Our findings have implications for drug development, as they highlight the potential benefits and risks of pursuing cross-disease drug targets.
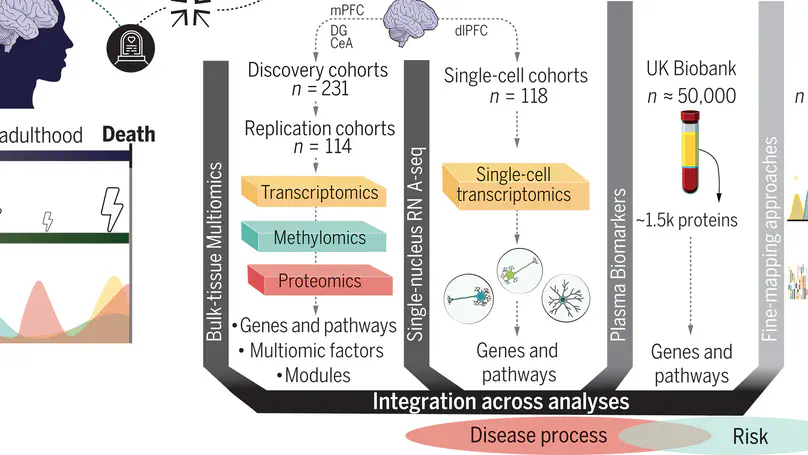
The molecular pathology of stress-related disorders remains elusive. Our brain multiregion, multiomic study of posttraumatic stress disorder (PTSD) and major depressive disorder (MDD) included the central nucleus of the amygdala, hippocampal dentate gyrus, and medial prefrontal cortex (mPFC). Genes and exons within the mPFC carried most disease signals replicated across two independent cohorts. Pathways pointed to immune function, neuronal and synaptic regulation, and stress hormones. Multiomic factor and gene network analyses provided the underlying genomic structure. Single nucleus RNA sequencing in dorsolateral PFC revealed dysregulated (stress-related) signals in neuronal and non-neuronal cell types. Analyses of brain-blood intersections in >50,000 UK Biobank participants were conducted along with fine-mapping of the results of PTSD and MDD genome-wide association studies to distinguish risk from disease processes. Our data suggest shared and distinct molecular pathology in both disorders and propose potential therapeutic targets and biomarkers.
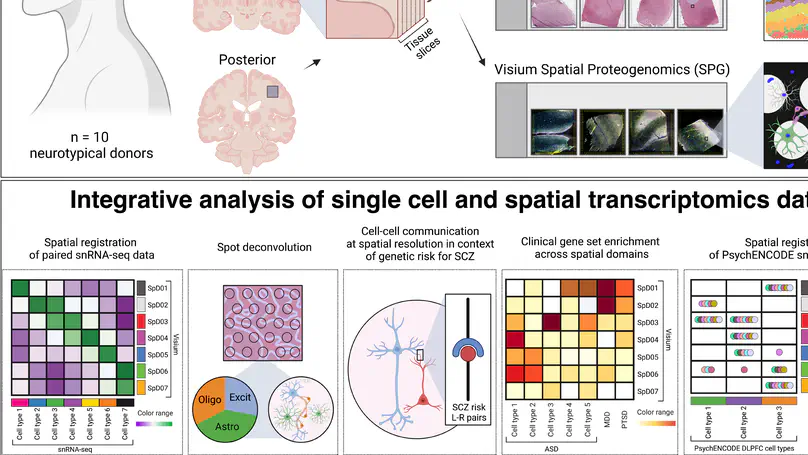
The molecular organization of the human neocortex historically has been studied in the context of its histological layers. However, emerging spatial transcriptomic technologies have enabled unbiased identification of transcriptionally defined spatial domains that move beyond classic cytoarchitecture. We used the Visium spatial gene expression platform to generate a data-driven molecular neuroanatomical atlas across the anterior-posterior axis of the human dorsolateral prefrontal cortex. Integration with paired single-nucleus RNA-sequencing data revealed distinct cell type compositions and cell-cell interactions across spatial domains. Using PsychENCODE and publicly available data, we mapped the enrichment of cell types and genes associated with neuropsychiatric disorders to discrete spatial domains.
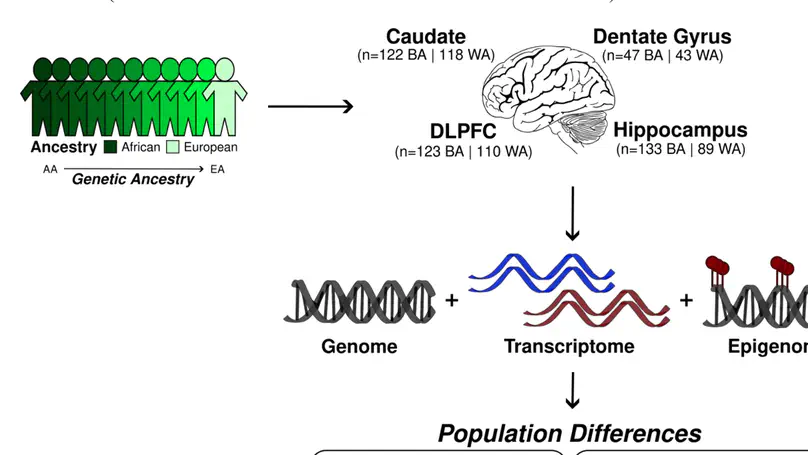
Ancestral differences in genomic variation affect the regulation of gene expression; however, most gene expression studies have been limited to European ancestry samples or adjusted to identify ancestry-independent associations. Here, we instead examined the impact of genetic ancestry on gene expression and DNA methylation in the postmortem brain tissue of admixed Black American neurotypical individuals to identify ancestry-dependent and ancestry-independent contributions. Ancestry-associated differentially expressed genes (DEGs), transcripts and gene networks, while notably not implicating neurons, are enriched for genes related to the immune response and vascular tissue and explain up to 26% of heritability for ischemic stroke, 27% of heritability for Parkinson disease and 30% of heritability for Alzheimer’s disease. Ancestry-associated DEGs also show general enrichment for the heritability of diverse immune-related traits but depletion for psychiatric-related traits. We also compared Black and non-Hispanic white Americans, confirming most ancestry-associated DEGs. Our results delineate the extent to which genetic ancestry affects differences in gene expression in the human brain and the implications for brain illness risk.
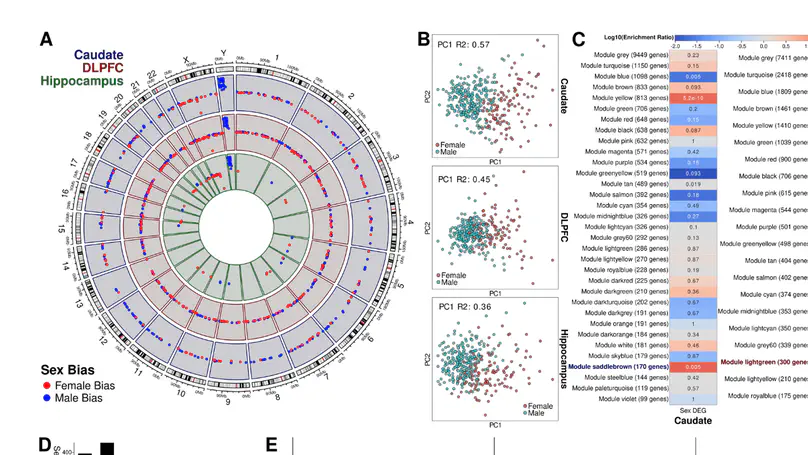
Schizophrenia is a complex neuropsychiatric disorder with sexually dimorphic features, including differential symptomatology, drug responsiveness, and male incidence rate. Prior large-scale transcriptome analyses for sex differences in schizophrenia have focused on the prefrontal cortex. Analyzing BrainSeq Consortium data (caudate nucleus: n = 399, dorsolateral prefrontal cortex: n = 377, and hippocampus: n = 394), we identified 831 unique genes that exhibit sex differences across brain regions, enriched for immune-related pathways. We observed X-chromosome dosage reduction in the hippocampus of male individuals with schizophrenia. Our sex interaction model revealed 148 junctions dysregulated in a sex-specific manner in schizophrenia. Sex-specific schizophrenia analysis identified dozens of differentially expressed genes, notably enriched in immune-related pathways. Finally, our sex-interacting expression quantitative trait loci analysis revealed 704 unique genes, nine associated with schizophrenia risk. These findings emphasize the importance of sex-informed analysis of sexually dimorphic traits, inform personalized therapeutic strategies in schizophrenia, and highlight the need for increased female samples for schizophrenia analyses.
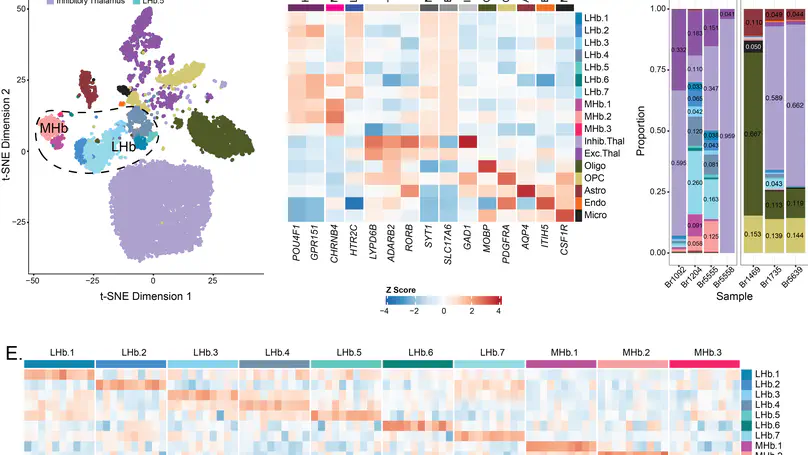
Pathophysiology of many neuropsychiatric disorders, including schizophrenia (SCZD), is linked to habenula (Hb) function. While pharmacotherapies and deep brain stimulation targeting the Hb are emerging as promising therapeutic treatments, little is known about the cell type-specific transcriptomic organization of the human Hb or how it is altered in SCZD. Here we define the molecular neuroanatomy of the human Hb and identify transcriptomic changes in individuals with SCZD compared to neurotypical controls. Utilizing Hb-enriched postmortem human brain tissue, we performed single nucleus RNA-sequencing (snRNA-seq; n=7 neurotypical donors) and identified 17 molecularly defined Hb cell types across 16,437 nuclei, including 3 medial and 7 lateral Hb populations, several of which were conserved between rodents and humans. Single molecule fluorescent in situ hybridization (smFISH; n=3 neurotypical donors) validated snRNA-seq Hb cell types and mapped their spatial locations. Bulk RNA-sequencing and cell type deconvolution in Hb-enriched tissue from 35 individuals with SCZD and 33 neurotypical controls yielded 45 SCZD-associated differentially expressed genes (DEGs, FDR < 0.05), with 32 (71%) unique to Hb-enriched tissue. eQTL analysis identified 717 independent SNP-gene pairs (FDR < 0.05), where either the SNP is a SCZD risk variant (16 pairs) or the gene is a SCZD DEG (7 pairs). eQTL and SCZD risk colocalization analysis identified 16 colocalized genes. These results identify topographically organized cell types with distinct molecular signatures in the human Hb and demonstrate unique genetic changes associated with SCZD, thereby providing novel molecular insights into the role of Hb in neuropsychiatric disorders.
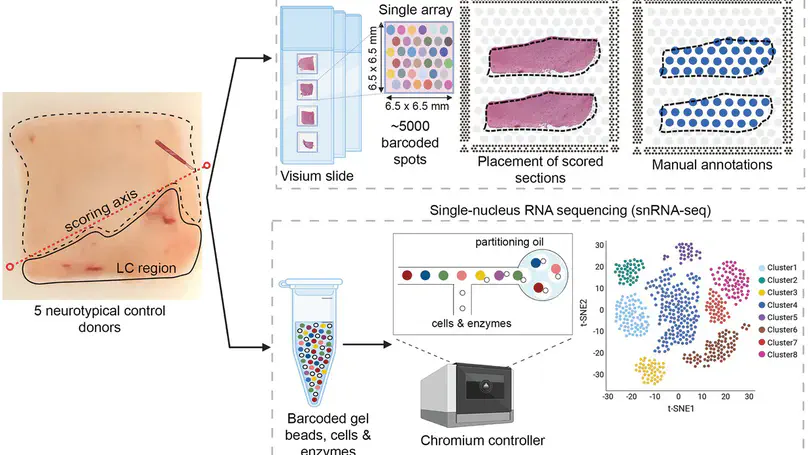
Norepinephrine (NE) neurons in the locus coeruleus (LC) make long-range projections throughout the central nervous system, playing critical roles in arousal and mood, as well as various components of cognition including attention, learning, and memory. The LC-NE system is also implicated in multiple neurological and neuropsychiatric disorders. Importantly, LC-NE neurons are highly sensitive to degeneration in both Alzheimer’s and Parkinson’s disease. Despite the clinical importance of the brain region and the prominent role of LC-NE neurons in a variety of brain and behavioral functions, a detailed molecular characterization of the LC is lacking. Here, we used a combination of spatially-resolved transcriptomics and single-nucleus RNA-sequencing to characterize the molecular landscape of the LC region and the transcriptomic profile of LC-NE neurons in the human brain. We provide a freely accessible resource of these data in web-accessible and downloadable formats.
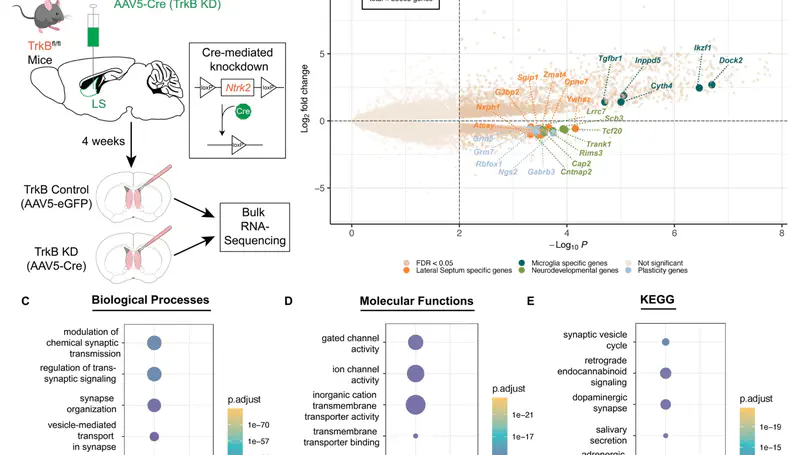
The lateral septum (LS), a GABAergic structure located in the basal forebrain, is implicated in social behavior, learning, and memory. We previously demonstrated that expression of tropomyosin kinase receptor B (TrkB) in LS neurons is required for social novelty recognition. To better understand molecular mechanisms by which TrkB signaling controls behavior, we locally knocked down TrkB in LS and used bulk RNA-sequencing to identify changes in gene expression downstream of TrkB. TrkB knockdown induces upregulation of genes associated with inflammation and immune responses, and downregulation of genes associated with synaptic signaling and plasticity. Next, we generated one of the first atlases of molecular profiles for LS cell types using single nucleus RNA-sequencing (snRNA-seq). We identified markers for the septum broadly, and the LS specifically, as well as for all neuronal cell types. We then investigated whether the differentially expressed genes (DEGs) induced by TrkB knockdown map to specific LS cell types. Enrichment testing identified that downregulated DEGs are broadly expressed across neuronal clusters. Enrichment analyses of these DEGs demonstrated that downregulated genes are uniquely expressed in the LS, and associated with either synaptic plasticity or neurodevelopmental disorders. Upregulated genes are enriched in LS microglia, associated with immune response and inflammation, and linked to both neurodegenerative disease and neuropsychiatric disorders. In addition, many of these genes are implicated in regulating social behaviors. In summary, the findings implicate TrkB signaling in the LS as a critical regulator of gene networks associated with psychiatric disorders that display social deficits, including schizophrenia and autism, and with neurodegenerative diseases, including Alzheimer’s.
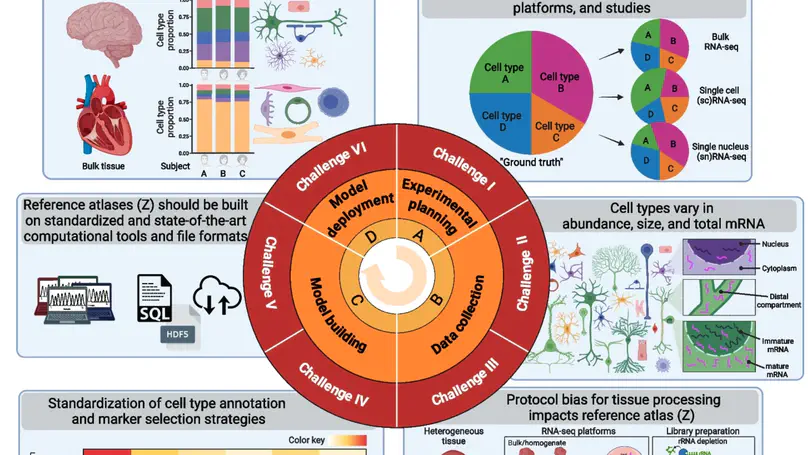
Deconvolution of cell mixtures in “bulk” transcriptomic samples from homogenate human tissue is important for understanding disease pathologies. However, several experimental and computational challenges impede transcriptomics-based deconvolution approaches using single-cell/nucleus RNA-seq reference atlases. Cells from the brain and blood have substantially different sizes, total mRNA, and transcriptional activities, and existing approaches may quantify total mRNA instead of cell type proportions. Further, standards are lacking for the use of cell reference atlases and integrative analyses of single-cell and spatial transcriptomics data. We discuss how to approach these key challenges with orthogonal “gold standard” datasets for evaluating deconvolution methods.
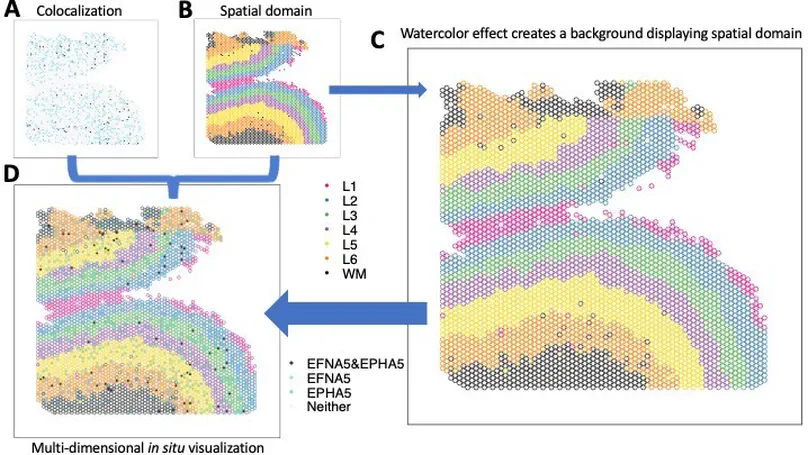
The creation of effective visualizations is a fundamental component of data analysis. In biomedical research, new challenges are emerging to visualize multi-dimensional data in a 2D space, but current data visualization tools have limited capabilities. To address this problem, we leverage Gestalt principles to improve the design and interpretability of multi-dimensional data in 2D data visualizations, layering aesthetics to display multiple variables. The proposed visualization can be applied to spatially-resolved transcriptomics data, but also broadly to data visualized in 2D space, such as embedding visualizations. We provide an open source R package escheR, which is built off of the state-of-the-art ggplot2 visualization framework and can be seamlessly integrated into genomics toolboxes and workflows.
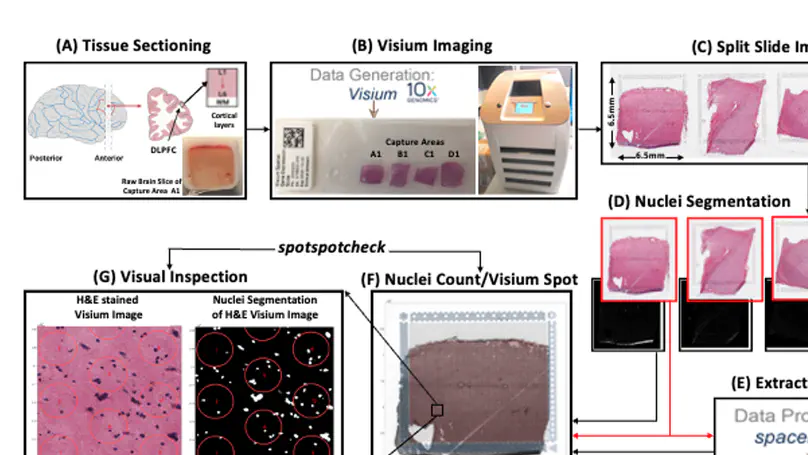
Spatially resolved transcriptomics (SRT) is a growing field that links gene expression to anatomical context. SRT approaches that use next-generation sequencing (NGS) combine RNA sequencing with histological or fluorescent imaging to generate spatial maps of gene expression in intact tissue sections. These technologies directly couple gene expression measurements with high-resolution histological or immunofluorescent images that contain rich morphological information about the tissue under study. While broad access to NGS-based spatial transcriptomic technology is now commercially available through the Visium platform from the vendor 10× Genomics, computational tools for extracting image-derived metrics for integration with gene expression data remain limited. We developed VistoSeg as a MATLAB pipeline to process, analyze and interactively visualize the high-resolution images generated in the Visium platform. VistoSeg outputs can be easily integrated with accompanying transcriptomic data to facilitate downstream analyses in common programing languages including R and Python. VistoSeg provides user-friendly tools for integrating image-derived metrics from histological and immunofluorescent images with spatially resolved gene expression data. Integration of this data enhances the ability to understand the transcriptional landscape within tissue architecture. VistoSeg is freely available at http://research.libd.org/VistoSeg/.
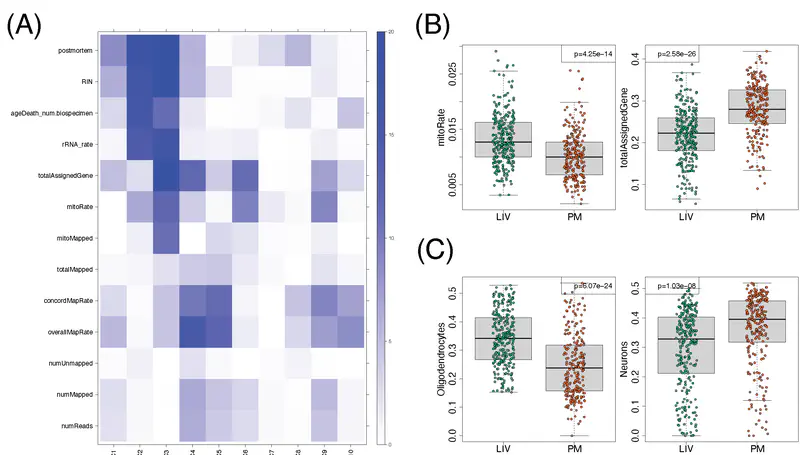
Molecular mechanisms of neuropsychiatric disorders are challenging to study in human brain. For decades, the preferred model has been to study postmortem human brain samples despite the limitations they entail. A recent study generated RNA sequencing data from biopsies of prefrontal cortex from living patients with Parkinson’s Disease and compared gene expression to postmortem tissue samples, from which they found vast differences between the two. This led the authors to question the utility of postmortem human brain studies. Through re-analysis of the same data, we unexpectedly found that the living brain tissue samples were of much lower quality than the postmortem samples across multiple standard metrics. We also performed simulations that illustrate the effects of ignoring RNA degradation in differential gene expression analyses, showing the effects can be substantial and of similar magnitude to what the authors find. For these reasons, we believe the authors’ conclusions are unjustified. To the contrary, while opportunities to study gene expression in the living brain are welcome, evidence that this eclipses the value of postmortem analyses is not apparent.
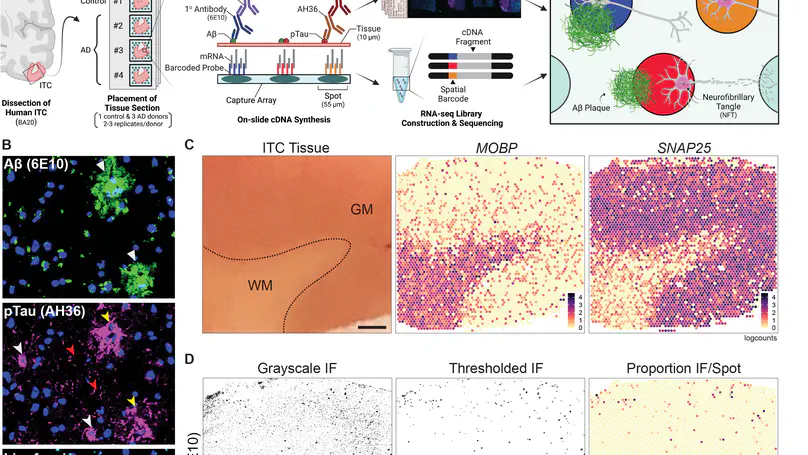
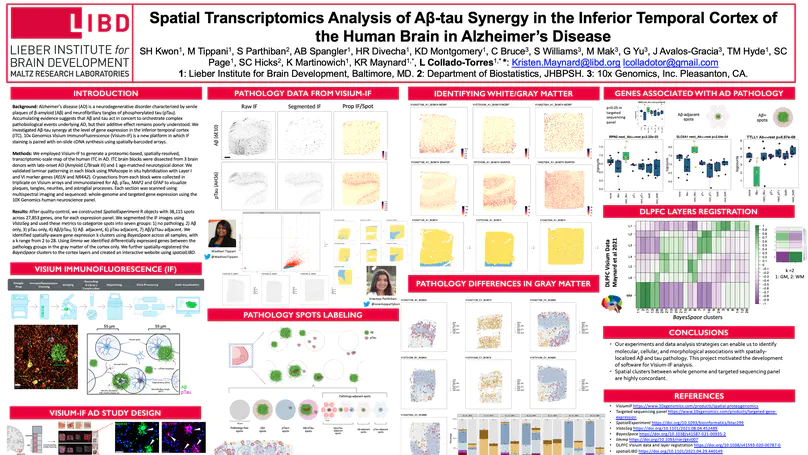
Neuropathological lesions in the brains of individuals affected with neurodegenerative disorders are hypothesized to trigger molecular and cellular processes that disturb the homeostasis of local microenvironments. Here, we applied the 10x Genomics Visium Spatial Proteogenomics (Visium-SPG) platform, which couples spatial gene expression with immunofluorescence (IF) protein co-detection, to evaluate its ability to quantify changes in spatial gene expression with respect to amyloid-beta (Aβ) and hyperphosphorylated tau (pTau) pathology in post-mortem human brain tissue from individuals with Alzheimer’s disease (AD). We identified transcriptomic signatures associated with proximity to Aβ in the human inferior temporal cortex during late-stage AD, which we further investigated at cellular resolution with combined IF and single-molecule fluorescent in situ hybridization (smFISH). The study provides a data analysis workflow for Visium-SPG, and the data represent a proof-of-principle for the power of multi-omic profiling in identifying changes in molecular dynamics that are spatially associated with pathology in the human brain. We provide the scientific community with web-based, interactive resources to access the datasets of the spatially resolved AD-related transcriptomes.
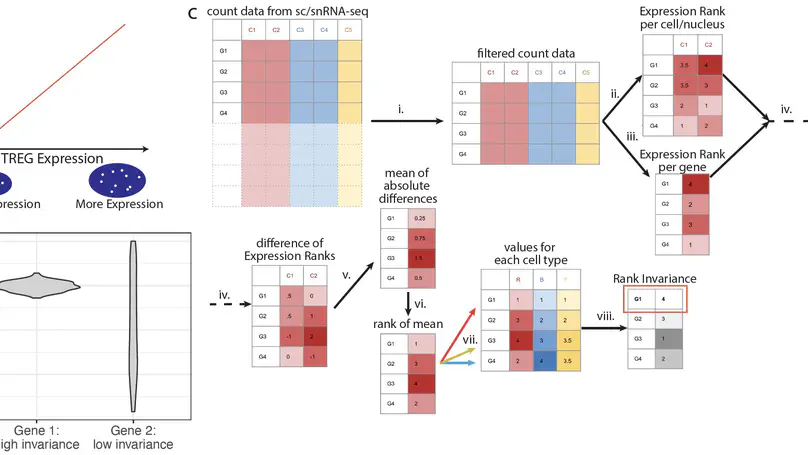
We define and identify a new class of control genes for next-generation sequencing called total RNA expression genes (TREGs), which correlate with total RNA abundance in cell types of different sizes and transcriptional activity. We provide a data-driven method to identify TREGs from single-cell RNA sequencing data, allowing the estimation of total amount of RNA when restricted to quantifying a limited number of genes. We demonstrate our method in postmortem human brain using multiplex single-molecule fluorescent in situ hybridization and compare candidate TREGs against classic housekeeping genes. We identify AKT3 as a top TREG across five brain regions.
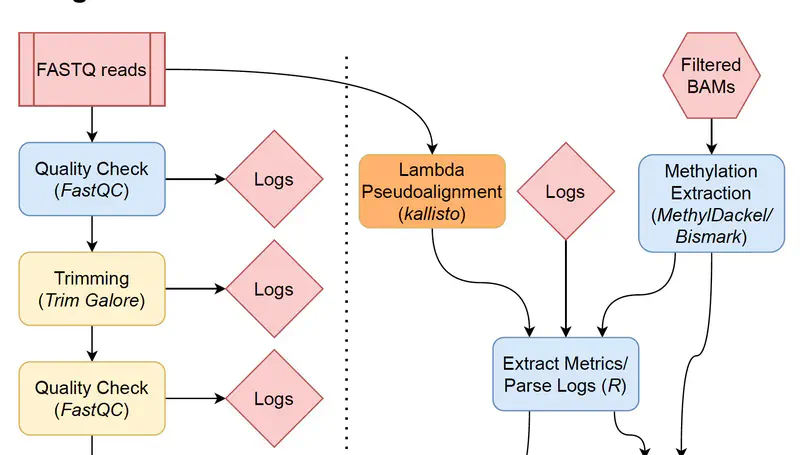
Background Bisulfite sequencing is a powerful tool for profiling genomic methylation, an epigenetic modification critical in the understanding of cancer, psychiatric disorders, and many other conditions. Raw data generated by whole genome bisulfite sequencing (WGBS) requires several computational steps before it is ready for statistical analysis, and particular care is required to process data in a timely and memory-efficient manner. Alignment to a reference genome is one of the most computationally demanding steps in a WGBS workflow, taking several hours or even days with commonly used WGBS-specific alignment software. This naturally motivates the creation of computational workflows that can utilize GPU-based alignment software to greatly speed up the bottleneck step. In addition, WGBS produces raw data that is large and often unwieldy; a lack of memory-efficient representation of data by existing pipelines renders WGBS impractical or impossible to many researchers. Results We present BiocMAP, a Bioconductor-friendly methylation analysis pipeline consisting of two modules, to address the above concerns. The first module performs computationally-intensive read alignment using Arioc, a GPU-accelerated short-read aligner. Since GPUs are not always available on the same computing environments where traditional CPU-based analyses are convenient, the second module may be run in a GPU-free environment. This module extracts and merges DNA methylation proportions—the fractions of methylated cytosines across all cells in a sample at a given genomic site. Bioconductor-based output objects in R utilize an on-disk data representation to drastically reduce required main memory and make WGBS projects computationally feasible to more researchers. Conclusions BiocMAP is implemented using Nextflow and available at http://research.libd.org/BiocMAP/. To enable reproducible analysis across a variety of typical computing environments, BiocMAP can be containerized with Docker or Singularity, and executed locally or with the SLURM or SGE scheduling engines. By providing Bioconductor objects, BiocMAP’s output can be integrated with powerful analytical open source software for analyzing methylation data.
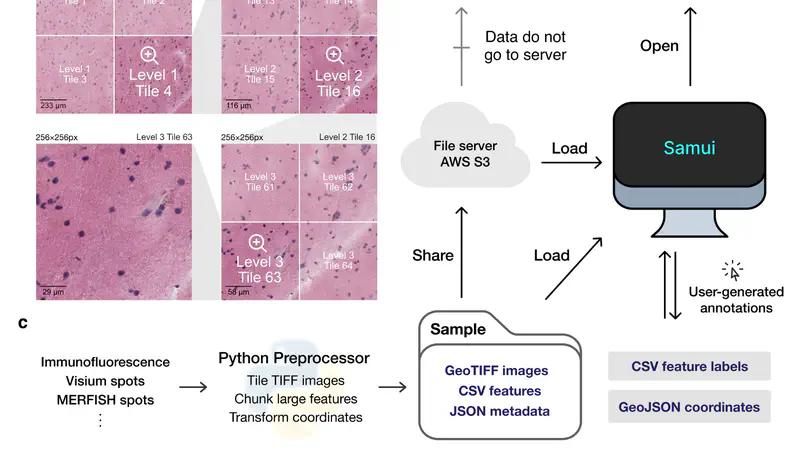
High-resolution and multiplexed imaging techniques are giving us an increasingly detailed observation of a biological system. However, sharing, exploring, and customizing the visualization of large multidimensional images can be a challenge. Here, we introduce Samui, a performant and interactive image visualization tool that runs completely in the web browser. Samui is specifically designed for fast image visualization and annotation and enables users to browse through large images and their selected features within seconds of receiving a link. We demonstrate the broad utility of Samui with images generated with two platforms: Vizgen MERFISH and 10x Genomics Visium Spatial Gene Expression. Samui along with example datasets is available at https://samuibrowser.com.
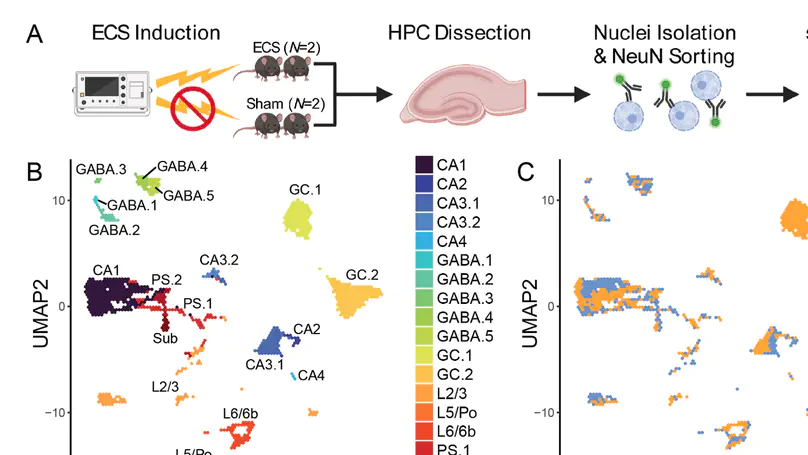
Activity-regulated gene (ARG) expression patterns in the hippocampus (HPC) regulate synaptic plasticity, learning, and memory, and are linked to both risk and treatment responses for many neuropsychiatric disorders. The HPC contains discrete classes of neurons with specialized functions, but cell type-specific activity-regulated transcriptional programs are not well characterized. Here, we used single-nucleus RNA-sequencing (snRNA-seq) in a mouse model of acute electroconvulsive seizures (ECS) to identify cell type-specific molecular signatures associated with induced activity in HPC neurons. We used unsupervised clustering and a priori marker genes to computationally annotate 15,990 high-quality HPC neuronal nuclei from N = 4 mice across all major HPC subregions and neuron types. Activity-induced transcriptomic responses were divergent across neuron populations, with dentate granule cells being particularly responsive to activity. Differential expression analysis identified both upregulated and downregulated cell type-specific gene sets in neurons following ECS. Within these gene sets, we identified enrichment of pathways associated with varying biological processes such as synapse organization, cellular signaling, and transcriptional regulation. Finally, we used matrix factorization to reveal continuous gene expression patterns differentially associated with cell type, ECS, and biological processes. This work provides a rich resource for interrogating activity-regulated transcriptional responses in HPC neurons at single-nuclei resolution in the context of ECS, which can provide biological insight into the roles of defined neuronal subtypes in HPC function.
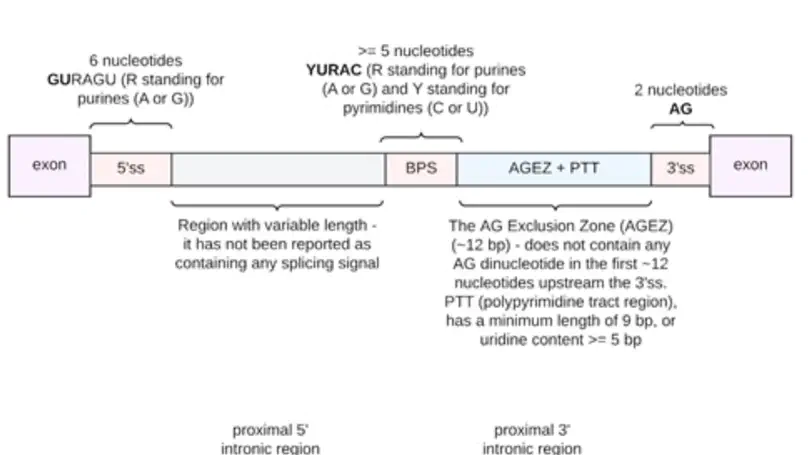
Dysregulation of RNA splicing contributes to both rare and complex diseases. RNA-sequencing data from human tissues has shown that this process can be inaccurate, resulting in the presence of novel introns detected at low frequency across samples and within an individual. To enable the full spectrum of intron use to be explored, we have developed IntroVerse, which offers an extensive catalogue on the splicing of 332,571 annotated introns and a linked set of 4,679,474 novel junctions covering 32,669 different genes. This dataset has been generated through the analysis of 17,510 human control RNA samples from 54 tissues provided by the Genotype-Tissue Expression Consortium. IntroVerse has two unique features: (i) it provides a complete catalogue of novel junctions and (ii) each novel junction has been assigned to a specific annotated intron. This unique, hierarchical structure offers multiple uses, including the identification of novel transcripts from known genes and their tissue-specific usage, and the assessment of background splicing noise for introns thought to be mis-spliced in disease states. IntroVerse provides a user-friendly web interface and is freely available at https://rytenlab.com/browser/app/introverse.
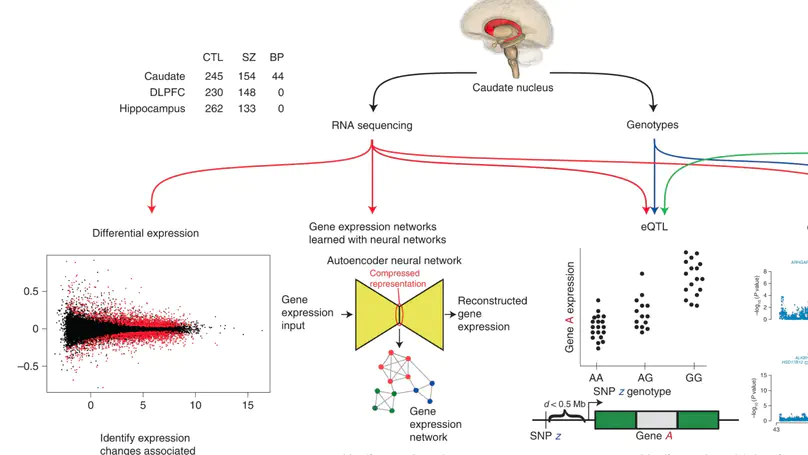
Most studies of gene expression in the brains of individuals with schizophrenia have focused on cortical regions, but subcortical nuclei such as the striatum are prominently implicated in the disease, and current antipsychotic drugs target the striatum’s dense dopaminergic innervation. Here, we performed a comprehensive analysis of the genetic and transcriptional landscape of schizophrenia in the postmortem caudate nucleus of the striatum of 443 individuals (245 neurotypical individuals, 154 individuals with schizophrenia and 44 individuals with bipolar disorder), 210 from African and 233 from European ancestries. Integrating expression quantitative trait loci analysis, Mendelian randomization with the latest schizophrenia genome-wide association study, transcriptome-wide association study and differential expression analysis, we identified many genes associated with schizophrenia risk, including potentially the dopamine D2 receptor short isoform. We found that antipsychotic medication has an extensive influence on caudate gene expression. We constructed caudate nucleus gene expression networks that highlight interactions involving schizophrenia risk. These analyses provide a resource for the study of schizophrenia and insights into risk mechanisms and potential therapeutic targets.
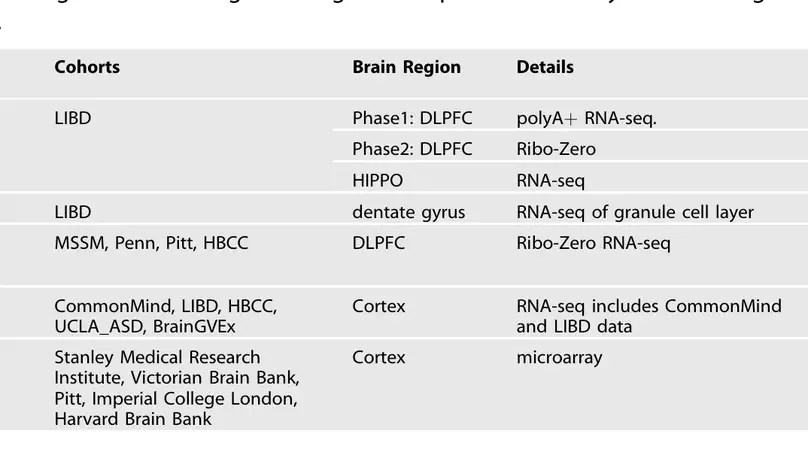
Following the recent review by Merikangas et al. summarizing differential gene expression in schizophrenia, we wanted to provide readers with our perspective on the state of the field in order to highlight current resources and ongoing challenges. We have led data collection and analysis of post mortem brains of control and schizophrenia samples across multiple large-scale cohorts, and we have provided data and results resources for each of these studies (Table 1).
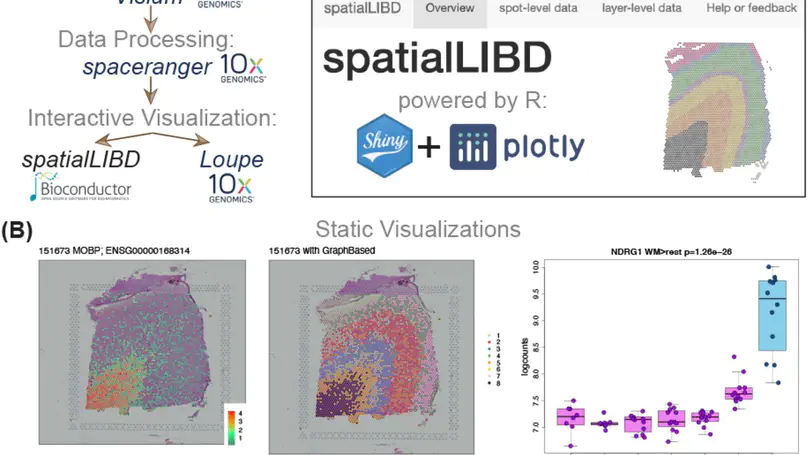
Background. Spatially-resolved transcriptomics has now enabled the quantification of high-throughput and transcriptome-wide gene expression in intact tissue while also retaining the spatial coordinates. Incorporating the precise spatial mapping of gene activity advances our understanding of intact tissue-specific biological processes. In order to interpret these novel spatial data types, interactive visualization tools are necessary. Results. We describe spatialLIBD, an R/Bioconductor package to interactively explore spatially-resolved transcriptomics data generated with the 10x Genomics Visium platform. The package contains functions to interactively access, visualize, and inspect the observed spatial gene expression data and data-driven clusters identified with supervised or unsupervised analyses, either on the user’s computer or through a web application. Conclusions. spatialLIBD is available at https://bioconductor.org/packages/spatialLIBD. It is fully compatible with SpatialExperiment and the Bioconductor ecosystem. Its functionality facilitates analyzing and interactively exploring spatially-resolved data from the Visium platform.
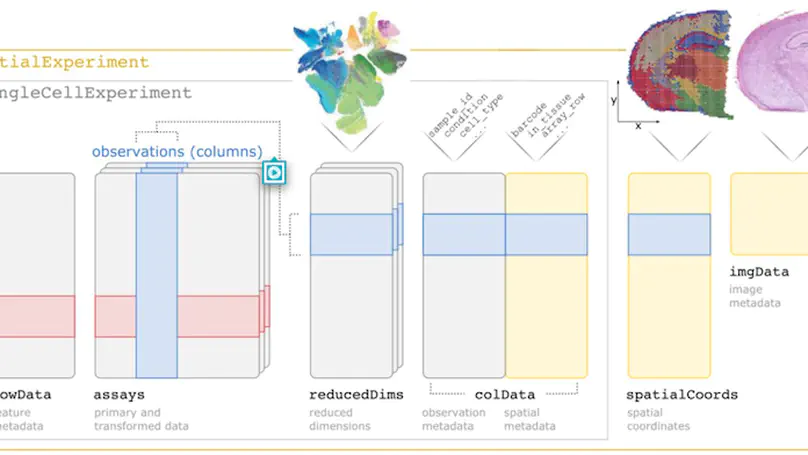
Summary SpatialExperiment is a new data infrastructure for storing and accessing spatially resolved transcriptomics data, implemented within the R/Bioconductor framework, which provides advantages of modularity, interoperability, standardized operations, and comprehensive documentation. Here, we demonstrate the structure and user interface with examples from the 10x Genomics Visium and seqFISH platforms, and provide access to example datasets and visualization tools in the STexampleData, TENxVisiumData, and ggspavis packages. Availability and Implementation The SpatialExperiment, STexampleData, TENxVisiumData, and ggspavis packages are available from Bioconductor. The package versions described in this manuscript are available in Bioconductor version 3.15 onwards. Supplementary information Supplementary tables and figures are available at Bioinformatics online.
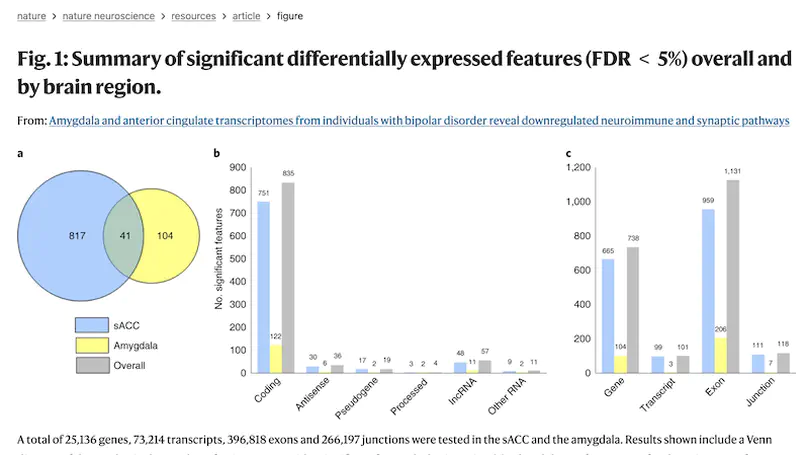
Recent genetic studies have identified variants associated with bipolar disorder (BD), but it remains unclear how brain gene expression is altered in BD and how genetic risk for BD may contribute to these alterations. Here, we obtained transcriptomes from subgenual anterior cingulate cortex and amygdala samples from post-mortem brains of individuals with BD and neurotypical controls, including 511 total samples from 295 unique donors. We examined differential gene expression between cases and controls and the transcriptional effects of BD-associated genetic variants. We found two coexpressed modules that were associated with transcriptional changes in BD: one enriched for immune and inflammatory genes and the other with genes related to the postsynaptic membrane. Over 50% of BD genome-wide significant loci contained significant expression quantitative trait loci (QTL) (eQTL), and these data converged on several individual genes, including SCN2A and GRIN2A. Thus, these data implicate specific genes and pathways that may contribute to the pathology of BD.
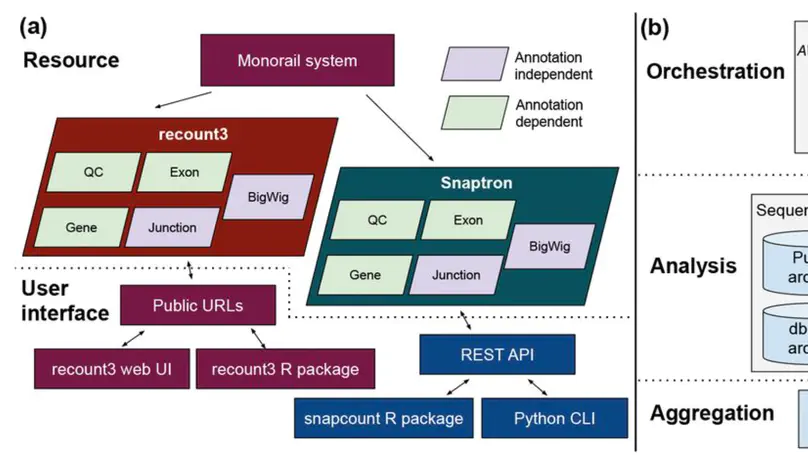
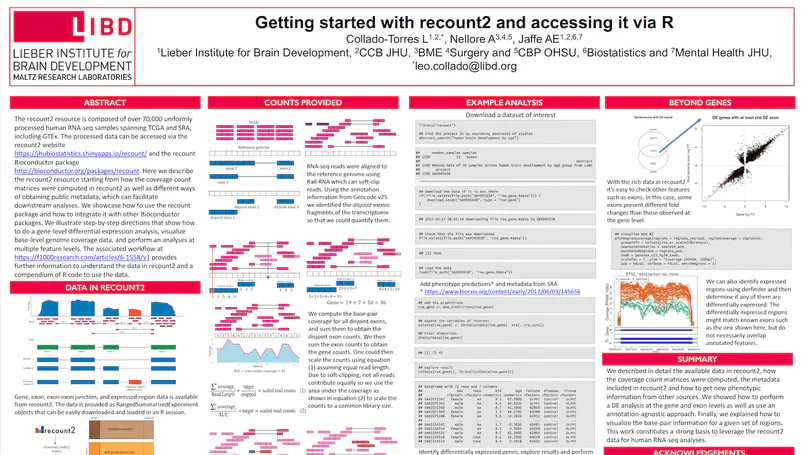
We present recount3, a resource consisting of over 750,000 publicly available human and mouse RNA sequencing (RNA-seq) samples uniformly processed by our new Monorail analysis pipeline. To facilitate access to the data, we provide the recount3 and snapcount R/Bioconductor packages as well as complementary web resources. Using these tools, data can be downloaded as study-level summaries or queried for specific exon-exon junctions, genes, samples, or other features. Monorail can be used to process local and/or private data, allowing results to be directly compared to any study in recount3. Taken together, our tools help biologists maximize the utility of publicly available RNA-seq data, especially to improve their understanding of newly collected data. recount3 is available from http://rna.recount.bio.
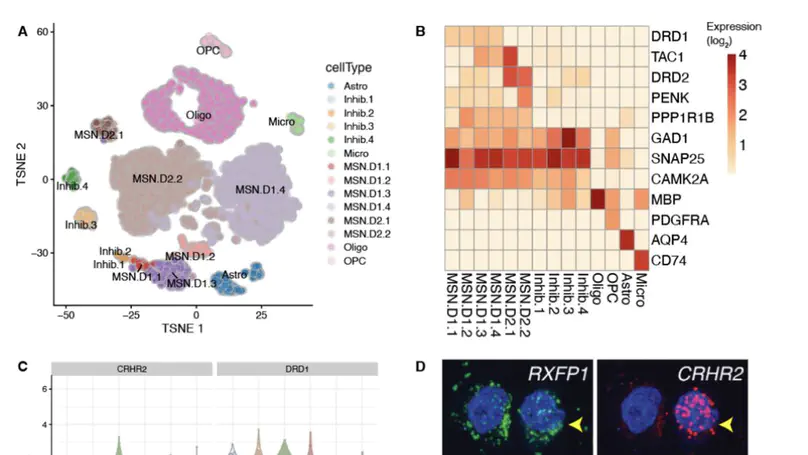
Single-cell gene expression technologies are powerful tools to study cell types in the human brain, but efforts have largely focused on cortical brain regions. We therefore created a single-nucleus RNA-sequencing resource of 70,615 high-quality nuclei to generate a molecular taxonomy of cell types across five human brain regions that serve as key nodes of the human brain reward circuitry: nucleus accumbens, amygdala, subgenual anterior cingulate cortex, hippocampus, and dorsolateral prefrontal cortex. We first identified novel subpopulations of interneurons and medium spiny neurons (MSNs) in the nucleus accumbens and further characterized robust GABAergic inhibitory cell populations in the amygdala. Joint analyses across the 107 reported cell classes revealed cell-type substructure and unique patterns of transcriptomic dynamics. We identified discrete subpopulations of D1- and D2-expressing MSNs in the nucleus accumbens to which we mapped cell-type-specific enrichment for genetic risk associated with both psychiatric disease and addiction.
DNA methylation (DNAm) is an epigenetic regulator of gene expression and a hallmark of gene-environment interaction. Using whole-genome bisulfite sequencing, we have surveyed DNAm in 344 samples of human postmortem brain tissue from neurotypical subjects and individuals with schizophrenia. We identify genetic influence on local methylation levels throughout the genome, both at CpG sites and CpH sites, with 86% of SNPs and 55% of CpGs being part of methylation quantitative trait loci (meQTLs). These associations can further be clustered into regions that are differentially methylated by a given SNP, highlighting the genes and regions with which these loci are epigenetically associated. These findings can be used to better characterize schizophrenia GWAS-identified variants as epigenetic risk variants. Regions differentially methylated by schizophrenia risk-SNPs explain much of the heritability associated with risk loci, despite covering only a fraction of the genomic space. We provide a comprehensive, single base resolution view of association between genetic variation and genomic methylation, and implicate schizophrenia GWAS-associated variants as influencing the epigenetic plasticity of the brain.
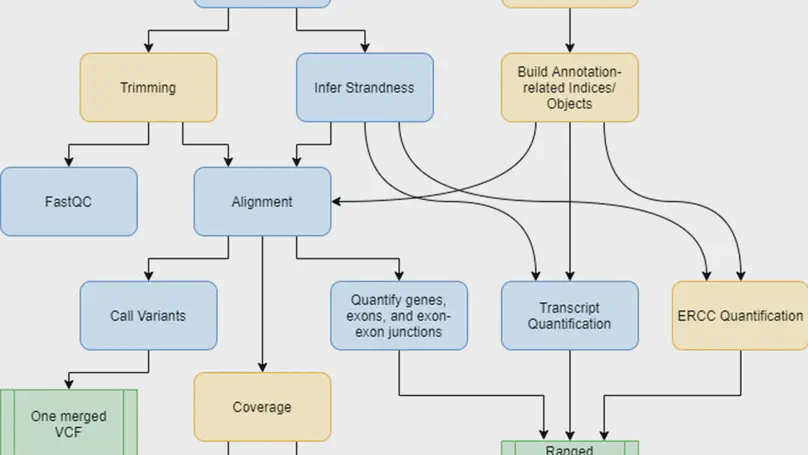
Background. RNA sequencing (RNA-seq) is a common and widespread biological assay, and an increasing amount of data is generated with it. In practice, there are a large number of individual steps a researcher must perform before raw RNA-seq reads yield directly valuable information, such as differential gene expression data. Existing software tools are typically specialized, only performing one step–such as alignment of reads to a reference genome–of a larger workflow. The demand for a more comprehensive and reproducible workflow has led to the production of a number of publicly available RNA-seq pipelines. However, we have found that most require computational expertise to set up or share among several users, are not actively maintained, or lack features we have found to be important in our own analyses. Results. In response to these concerns, we have developed a Scalable Pipeline for Expression Analysis and Quantification (SPEAQeasy), which is easy to install and share, and provides a bridge towards R/Bioconductor downstream analysis solutions. SPEAQeasy is portable across computational frameworks (SGE, SLURM, local, docker integration) and different configuration files are provided (http://research.libd.org/SPEAQeasy/). Conclusions. SPEAQeasy is user-friendly and lowers the computational-domain entry barrier for biologists and clinicians to RNA-seq data processing as the main input file is a table with sample names and their corresponding FASTQ files. The goal is to provide a flexible pipeline that is immediately usable by researchers, regardless of their technical background or computing environment.
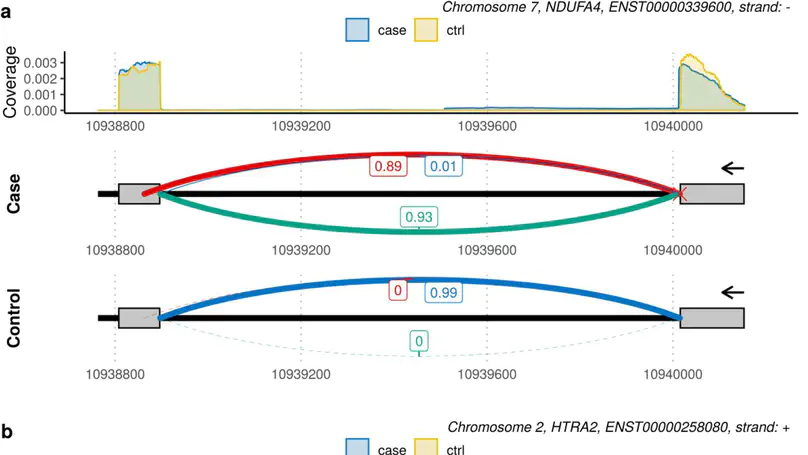
Although next-generation sequencing technologies have accelerated the discovery of novel gene-to-disease associations, many patients with suspected Mendelian diseases still leave the clinic without a genetic diagnosis. An estimated one third of these patients will have disorders caused by mutations impacting splicing. RNA-sequencing has been shown to be a promising diagnostic tool, however few methods have been developed to integrate RNA-sequencing data into the diagnostic pipeline. Here, we introduce dasper, an R/Bioconductor package that improves upon existing tools for detecting aberrant splicing by using machine learning to incorporate disruptions in exon-exon junction counts as well as coverage. dasper is designed for diagnostics, providing a rank-based report of how aberrant each splicing event looks, as well as including visualization functionality to facilitate interpretation. We validate dasper using 16 patient-derived fibroblast cell lines harbouring pathogenic variants known to impact splicing. We find that dasper is able to detect pathogenic splicing events with greater accuracy than existing LeafCutterMD or z-score approaches. Furthermore, by only applying a broad OMIM gene filter (without any variant-level filters), dasper is able to detect pathogenic splicing events within the top 10 most aberrant identified for each patient. Since using publicly available control data minimises costs associated with incorporating RNA-sequencing into diagnostic pipelines, we also investigate the use of 504 GTEx fibroblast samples as controls. We find that dasper leverages publicly available data effectively, ranking pathogenic splicing events in the top 25. Thus, we believe dasper can increase diagnostic yield for a pathogenic splicing variants and enable the efficient implementation of RNA-sequencing for diagnostics in clinical laboratories.
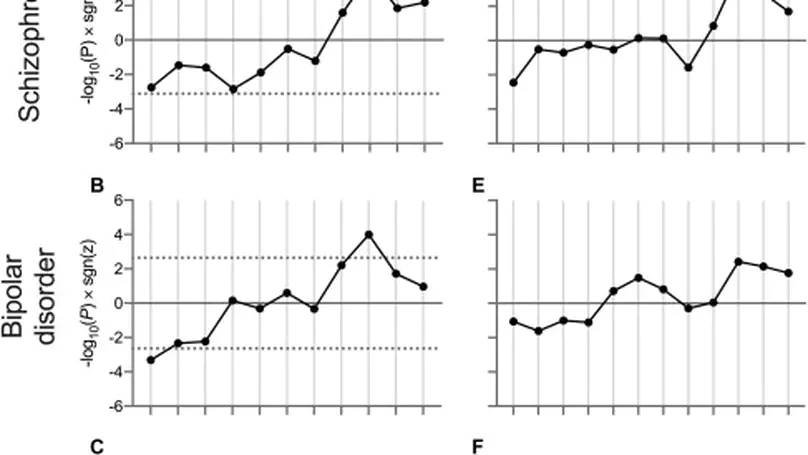
Background. Recent breakthroughs in psychiatric genetics have implicated biological pathways onto which genetic risk for psychiatric disorders converges. However, these studies do not reveal the developmental time point(s) at which these pathways are relevant. Methods. We aimed to determine the relationship between psychiatric risk and developmental gene expression relating to discrete biological pathways. We used postmortem RNA sequencing data (BrainSeq and BrainSpan) from brain tissue at multiple prenatal and postnatal time points, with summary statistics from recent genome-wide association studies of schizophrenia, bipolar disorder, and major depressive disorder. We prioritized gene sets for overall enrichment of association with each disorder and then tested the relationship between the association of their constituent genes with their relative expression at each developmental stage. Results. We observed relationships between the expression of genes involved in voltage-gated cation channel activity during early midfetal, adolescence, and early adulthood time points and association with schizophrenia and bipolar disorder, such that genes more strongly associated with these disorders had relatively low expression during early midfetal development and higher expression during adolescence and early adulthood. The relationship with schizophrenia was strongest for the subset of genes related to calcium channel activity, while for bipolar disorder, the relationship was distributed between calcium and potassium channel activity genes. Conclusions. Our results indicate periods during development when biological pathways related to the activity of calcium and potassium channels may be most vulnerable to the effects of genetic variants conferring risk for psychiatric disorders. Furthermore, they indicate key time points and potential targets for disorder-specific therapeutic interventions.
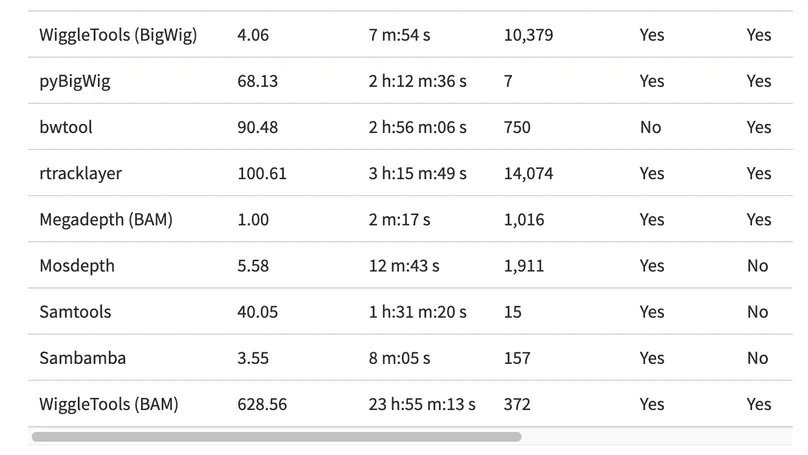
Motivation. A common way to summarize sequencing datasets is to quantify data lying within genes or other genomic intervals. This can be slow and can require different tools for different input file types. Results. Megadepth is a fast tool for quantifying alignments and coverage for BigWig and BAM/CRAM input files, using substantially less memory than the next-fastest competitor. Megadepth can summarize coverage within all disjoint intervals of the Gencode V35 gene annotation for more than 19 000 GTExV8 BigWig files in approximately 1 h using 32 threads. Megadepth is available both as a command-line tool and as an R/Bioconductor package providing much faster quantification compared to the rtracklayer package. Availability and implementation: https://github.com/ChristopherWilks/megadepth, https://bioconductor.org/packages/megadepth.
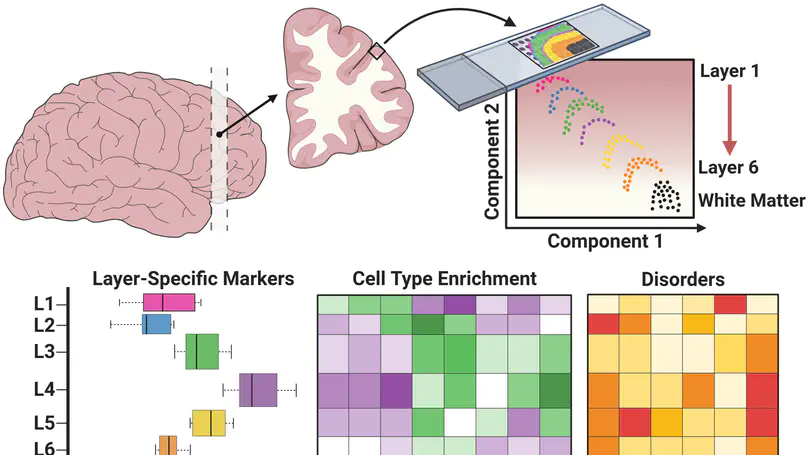
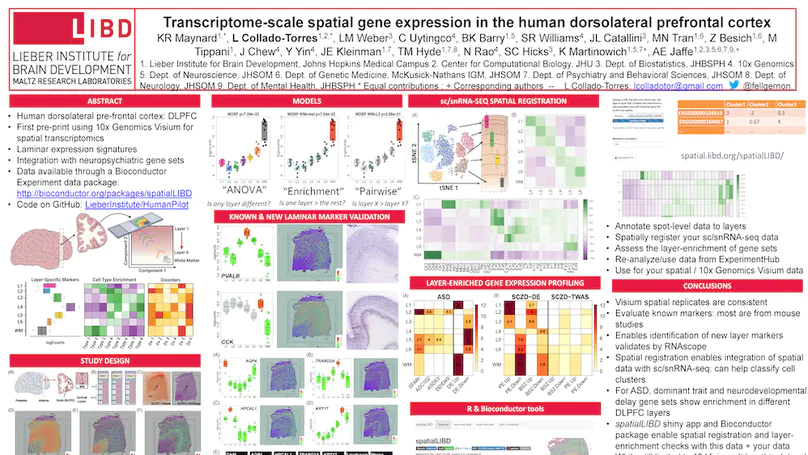
We used the 10x Genomics Visium platform to define the spatial topography of gene expression in the six-layered human dorsolateral prefrontal cortex. We identified extensive layer-enriched expression signatures and refined associations to previous laminar markers. We overlaid our laminar expression signatures on large-scale single nucleus RNA-sequencing data, enhancing spatial annotation of expression-driven clusters. By integrating neuropsychiatric disorder gene sets, we showed differential layer-enriched expression of genes associated with schizophrenia and autism spectrum disorder, highlighting the clinical relevance of spatially defined expression. We then developed a data-driven framework to define unsupervised clusters in spatial transcriptomics data, which can be applied to other tissues or brain regions in which morphological architecture is not as well defined as cortical laminae. Last, we created a web application for the scientific community to explore these raw and summarized data to augment ongoing neuroscience and spatial transcriptomics research (http://research.libd.org/spatialLIBD).
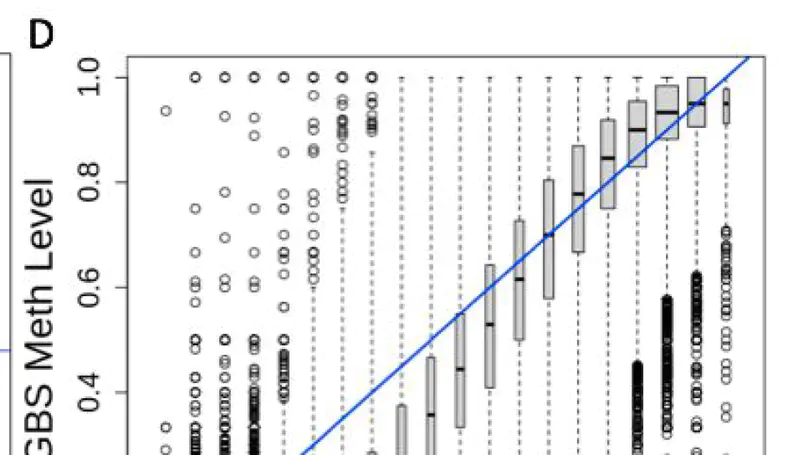
DNA methylation (DNAm) is a key epigenetic regulator of gene expression across development. The developing prenatal brain is a highly dynamic tissue, but our understanding of key drivers of epigenetic variability across development is limited. We, therefore, assessed genomic methylation at over 39 million sites in the prenatal cortex using whole-genome bisulfite sequencing and found loci and regions in which methylation levels are dynamic across development. We saw that DNAm at these loci was associated with nearby gene expression and enriched for enhancer chromatin states in prenatal brain tissue. Additionally, these loci were enriched for genes associated with neuropsychiatric disorders and genes involved with neurogenesis. We also found autosomal differences in DNAm between the sexes during prenatal development, though these have less clear functional consequences. We lastly confirmed that the dynamic methylation at this critical period is specifically CpG methylation, with generally low levels of CpH methylation. Our findings provide detailed insight into prenatal brain development as well as clues to the pathogenesis of psychiatric traits seen later in life.
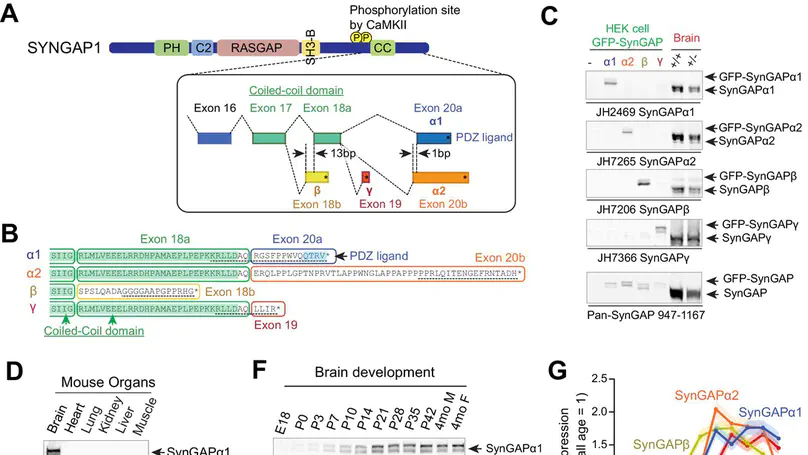
SynGAP is a synaptic Ras GTPase-activating protein (GAP) with four C-terminal splice variants: α1, α2, β, and γ. Although studies have implicated SYNGAP1 in several cognitive disorders, it is not clear which SynGAP isoforms contribute to disease. Here, we demonstrate that SynGAP isoforms exhibit unique spatiotemporal expression patterns and play distinct roles in neuronal and synaptic development in mouse neurons. SynGAP-α1, which undergoes liquid-liquid phase separation with PSD-95, is highly enriched in synapses and is required for LTP. In contrast, SynGAP-β, which does not bind PSD-95 PDZ domains, is less synaptically targeted and promotes dendritic arborization. A mutation in SynGAP-α1 that disrupts phase separation and synaptic targeting abolishes its ability to regulate plasticity and instead causes it to drive dendritic development like SynGAP-β. These results demonstrate that distinct intrinsic biochemical properties of SynGAP isoforms determine their function, and individual isoforms may differentially contribute to the pathogenesis of SYNGAP1-related cognitive disorders.
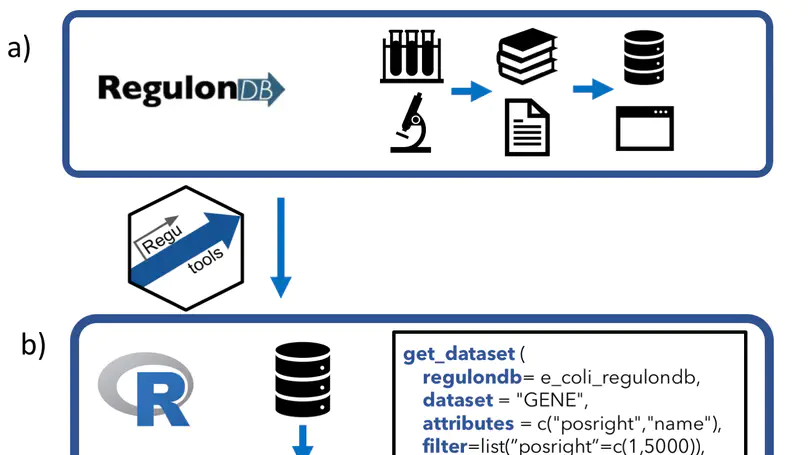
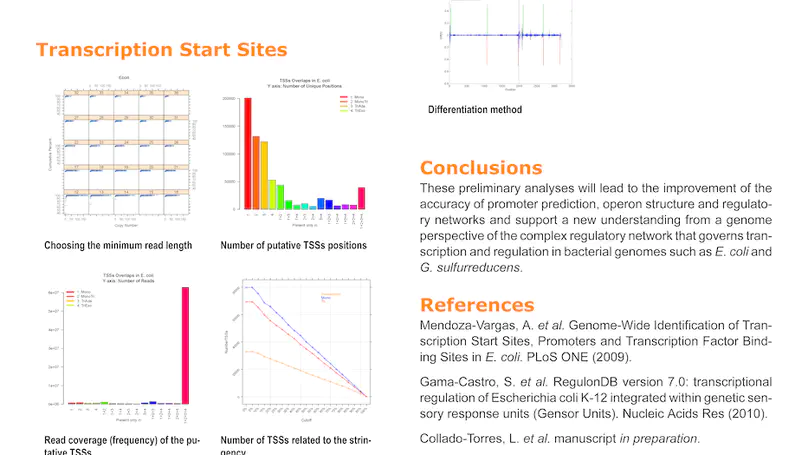
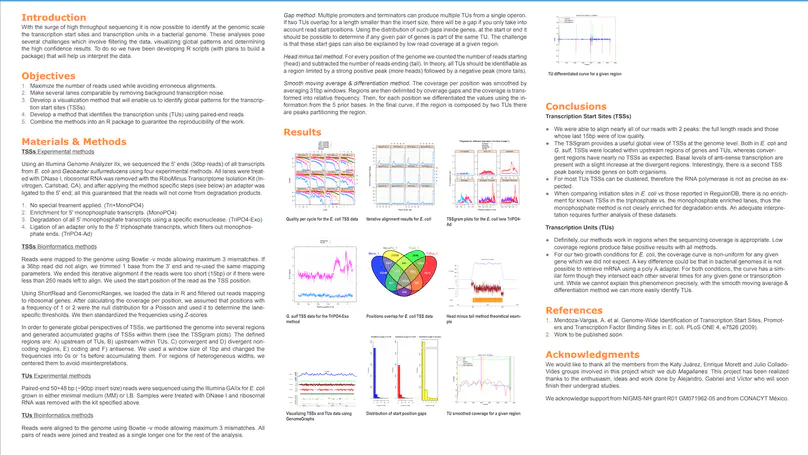
RegulonDB has collected, harmonized and centralized data from hundreds of experiments for nearly two decades and is considered a point of reference for transcriptional regulation in Escherichia coli K12. Here, we present the regutools R package to facilitate programmatic access to RegulonDB data in computational biology. regutools gives researchers the possibility of writing reproducible workflows with automated queries to RegulonDB. The regutools package serves as a bridge between RegulonDB data and the Bioconductor ecosystem by reusing the data structures and statistical methods powered by other Bioconductor packages. We demonstrate the integration of regutools with Bioconductor by analyzing transcription factor DNA binding sites and transcriptional regulatory networks from RegulonDB. We anticipate that regutools will serve as a useful building block in our progress to further our understanding of gene regulatory networks.
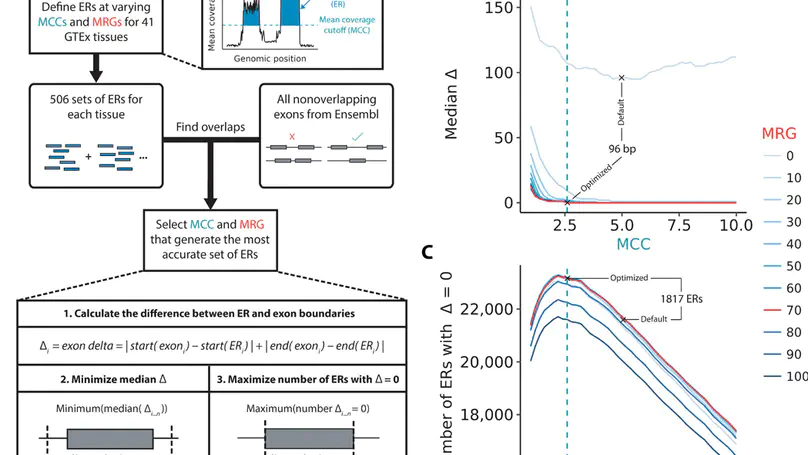
Growing evidence suggests that human gene annotation remains incomplete; however, it is unclear how this affects different tissues and our understanding of different disorders. Here, we detect previously unannotated transcription from Genotype-Tissue Expression RNA sequencing data across 41 human tissues. We connect this unannotated transcription to known genes, confirming that human gene annotation remains incomplete, even among well-studied genes including 63% of the Online Mendelian Inheritance in Man–morbid catalog and 317 neurodegeneration-associated genes. We find the greatest abundance of unannotated transcription in brain and genes highly expressed in brain are more likely to be reannotated. We explore examples of reannotated disease genes, such as SNCA, for which we experimentally validate a previously unidentified, brain-specific, potentially protein-coding exon. We release all tissue-specific transcriptomes through vizER: http://rytenlab.com/browser/app/vizER. We anticipate that this resource will facilitate more accurate genetic analysis, with the greatest impact on our understanding of Mendelian and complex neurogenetic disorders.
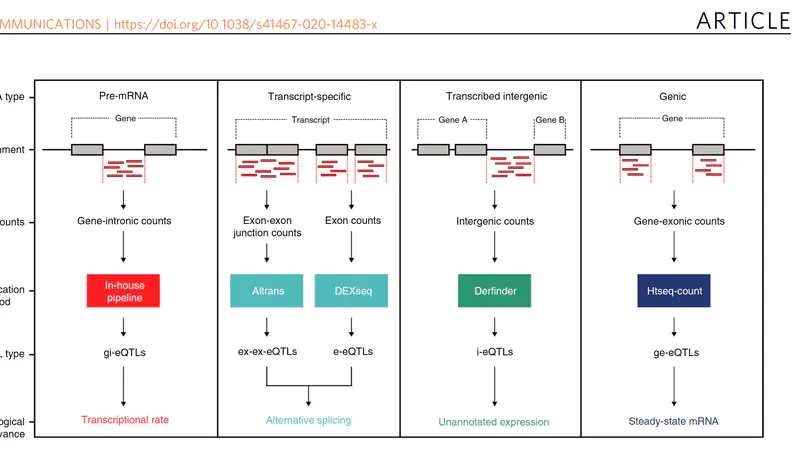
Genome-wide association studies have generated an increasing number of common genetic variants associated with neurological and psychiatric disease risk. An improved understanding of the genetic control of gene expression in human brain is vital considering this is the likely modus operandum for many causal variants. However, human brain sampling complexities limit the explanatory power of brain-related expression quantitative trait loci (eQTL) and allele-specific expression (ASE) signals. We address this, using paired genomic and transcriptomic data from putamen and substantia nigra from 117 human brains, interrogating regulation at different RNA processing stages and uncovering novel transcripts. We identify disease-relevant regulatory loci, find that splicing eQTLs are enriched for regulatory information of neuron-specific genes, that ASEs provide cell-specific regulatory information with evidence for cellular specificity, and that incomplete annotation of the brain transcriptome limits interpretation of risk loci for neuropsychiatric disease. This resource of regulatory data is accessible through our web server, http://braineacv2.inf.um.es/.
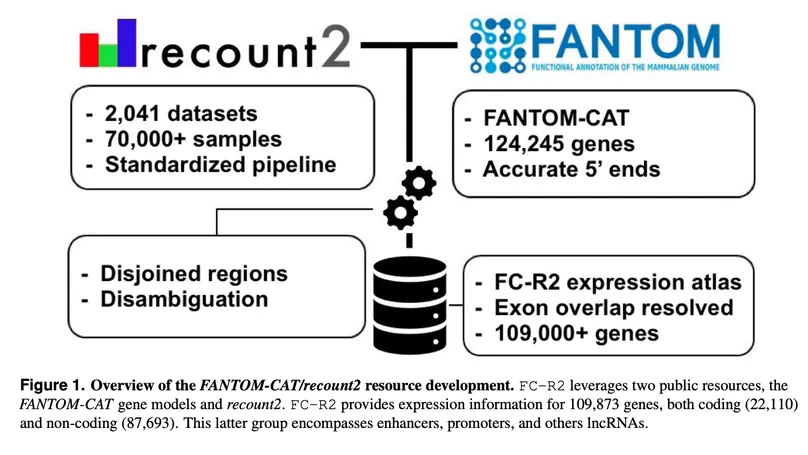
Long noncoding RNAs (lncRNAs) have emerged as key coordinators of biological and cellular processes. Characterizing lncRNA expression across cells and tissues is key to understanding their role in determining phenotypes including human diseases. We present here FC-R2, a comprehensive expression atlas across a broadly defined human transcriptome, inclusive of over 109,000 coding and noncoding genes, as described in the FANTOM CAGE-Associated Transcriptome (FANTOM-CAT) study. This atlas greatly extends the gene annotation used in the original recount2 resource. We demonstrate the utility of the FC-R2 atlas by reproducing key findings from published large studies and by generating new results across normal and diseased human samples. In particular, we (a) identify tissue-specific transcription profiles for distinct classes of coding and noncoding genes, (b) perform differential expression analysis across thirteen cancer types, identifying novel noncoding genes potentially involved in tumor pathogenesis and progression, and (c) confirm the prognostic value for several enhancers lncRNAs expression in cancer. Our resource is instrumental for the systematic molecular characterization of lncRNA by the FANTOM6 Consortium. In conclusion, comprised of over 70,000 samples, the FC-R2 atlas will empower other researchers to investigate functions and biological roles of both known coding genes and novel lncRNAs.
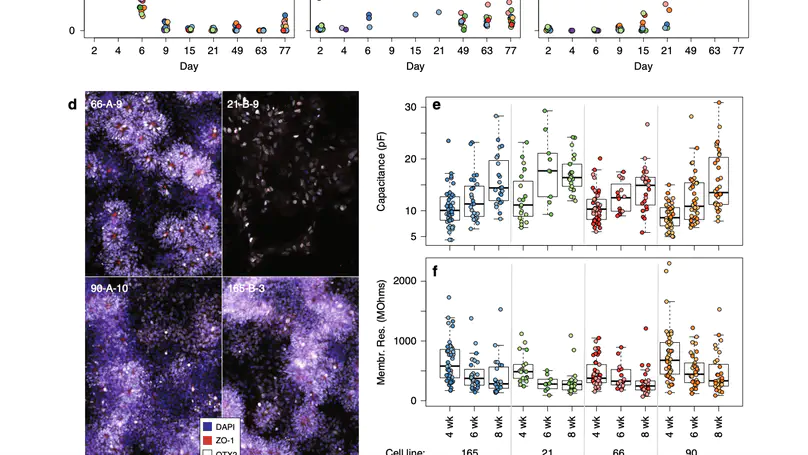
Human induced pluripotent stem cells (hiPSCs) are a powerful model of neural differentiation and maturation. We present a hiPSC transcriptomics resource on corticogenesis from 5 iPSC donor and 13 subclonal lines across 9 time points over 5 broad conditions: self-renewal, early neuronal differentiation, neural precursor cells (NPCs), assembled rosettes, and differentiated neuronal cells. We identify widespread changes in the expression of both individual features and global patterns of transcription. We next demonstrate that co-culturing human NPCs with rodent astrocytes results in mutually synergistic maturation, and that cell type-specific expression data can be extracted using only sequencing read alignments without cell sorting. We lastly adapt a previously generated RNA deconvolution approach to single-cell expression data to estimate the relative neuronal maturity of iPSC-derived neuronal cultures and human brain tissue. Using many public datasets, we demonstrate neuronal cultures are maturationally heterogeneous but contain subsets of neurons more mature than previously observed.
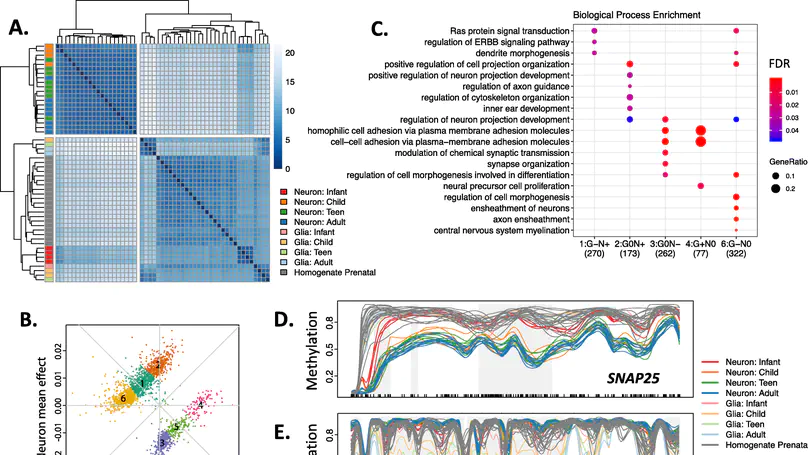
Background: DNA methylation (DNAm) is a critical regulator of both development and cellular identity and shows unique patterns in neurons. To better characterize maturational changes in DNAm patterns in these cells, we profile the DNAm landscape at single-base resolution across the first two decades of human neocortical development in NeuN+ neurons using whole-genome bisulfite sequencing and compare them to non-neurons (primarily glia) and prenatal homogenate cortex. Results: We show that DNAm changes more dramatically during the first 5 years of postnatal life than during the entire remaining period. We further refine global patterns of increasingly divergent neuronal CpG and CpH methylation (mCpG and mCpH) into six developmental trajectories and find that in contrast to genome-wide patterns, neighboring mCpG and mCpH levels within these regions are highly correlated. We integrate paired RNAseq data and identify putative regulation of hundreds of transcripts and their splicing events exclusively by mCpH levels, independently from mCpG levels, across this period. We finally explore the relationship between DNAm patterns and development of brain-related phenotypes and find enriched heritability for many phenotypes within identified DNAm features. Conclusions: By profiling DNAm changes in NeuN-sorted neurons over the span of human cortical development, we identify novel, dynamic regions of DNAm that would be masked in homogenate DNAm data; expand on the relationship between CpG methylation, CpH methylation, and gene expression; and find enrichment particularly for neuropsychiatric diseases in genomic regions with cell type-specific, developmentally dynamic DNAm patterns. Keywords: DNA methylation, Neurodevelopment, Gene expression, Non-CpG methylation.
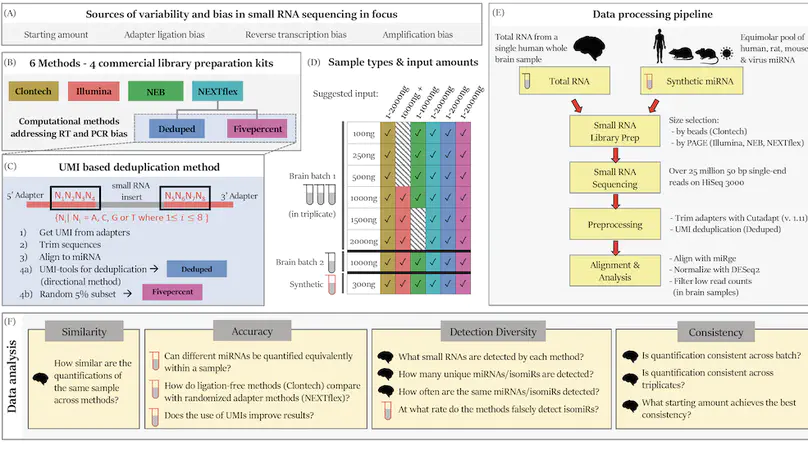
Background: RNA sequencing offers advantages over other quantification methods for microRNA (miRNA), yet numerous biases make reliable quantification challenging. Previous evaluations of these biases have focused on adapter ligation bias with limited evaluation of reverse transcription bias or amplification bias. Furthermore, evaluations of the quantification of isomiRs (miRNA isoforms) or the influence of starting amount on performance have been very limited. No study had yet evaluated the quantification of isomiRs of altered length or compared the consistency of results derived from multiple moderate starting inputs. We therefore evaluated quantifications of miRNA and isomiRs using four library preparation kits, with various starting amounts, as well as quantifications following removal of duplicate reads using unique molecular identifiers (UMIs) to mitigate reverse transcription and amplification biases. Results: All methods resulted in false isomiR detection; however, the adapter-free method tested was especially prone to false isomiR detection. We demonstrate that using UMIs improves accuracy and we provide a guide for input amounts to improve consistency. Conclusions: Our data show differences and limitations of current methods, thus raising concerns about the validity of quantification of miRNA and isomiRs across studies. We advocate for the use of UMIs to improve accuracy and reliability of miRNA quantifications.
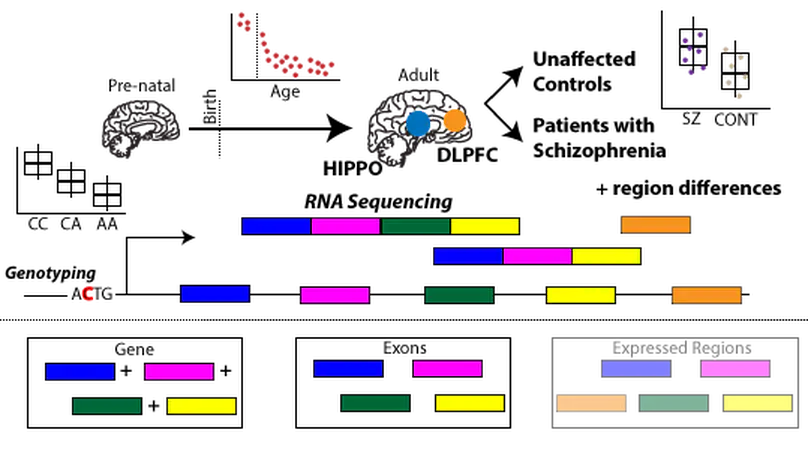
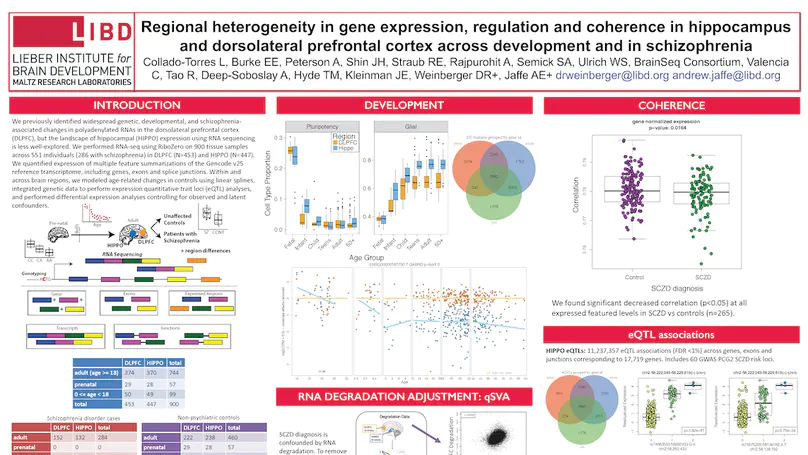
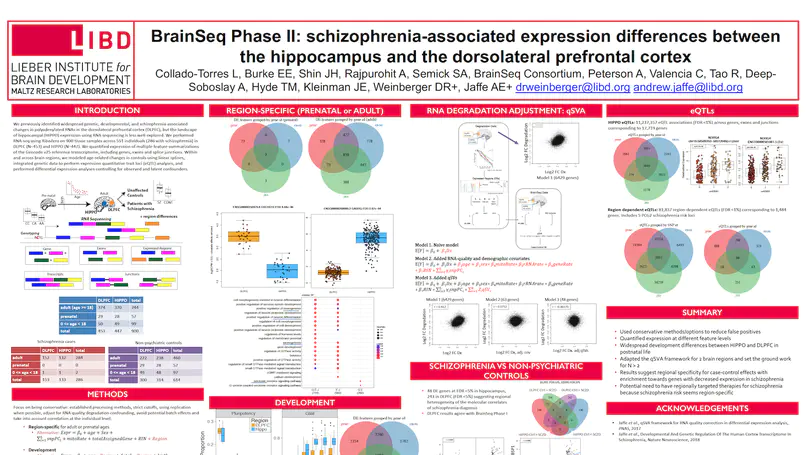
The hippocampus formation, although prominently implicated in schizophrenia pathogenesis, has been overlooked in large-scale genomics efforts in the schizophrenic brain. We performed RNA-seq in hippocampi and dorsolateral prefrontal cortices (DLPFCs) from 551 individuals (286 with schizophrenia). We identified substantial regional differences in gene expression and found widespread developmental differences that were independent of cellular composition. We identified 48 and 245 differentially expressed genes (DEGs) associated with schizophrenia within the hippocampus and DLPFC, with little overlap between the brain regions. 124 of 163 (76.6%) of schizophrenia GWAS risk loci contained eQTLs in any region. Transcriptome-wide association studies in each region identified many novel schizophrenia risk features that were brain region-specific. Last, we identified potential molecular correlates of in vivo evidence of altered prefrontal-hippocampal functional coherence in schizophrenia. These results underscore the complexity and regional heterogeneity of the transcriptional correlates of schizophrenia and offer new insights into potentially causative biology.
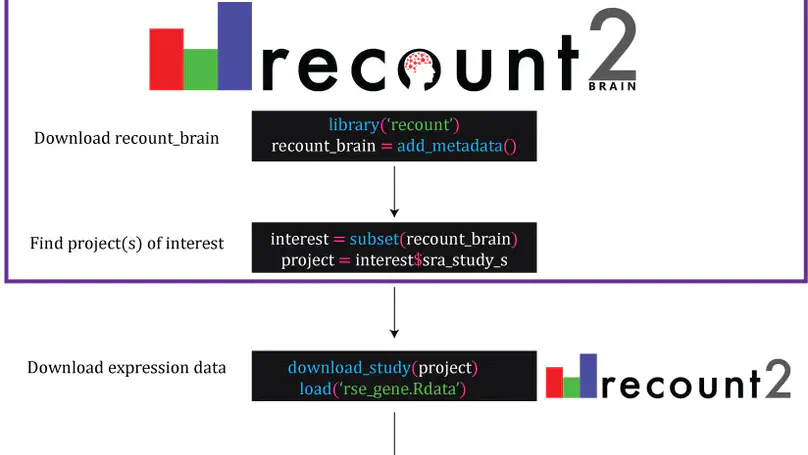
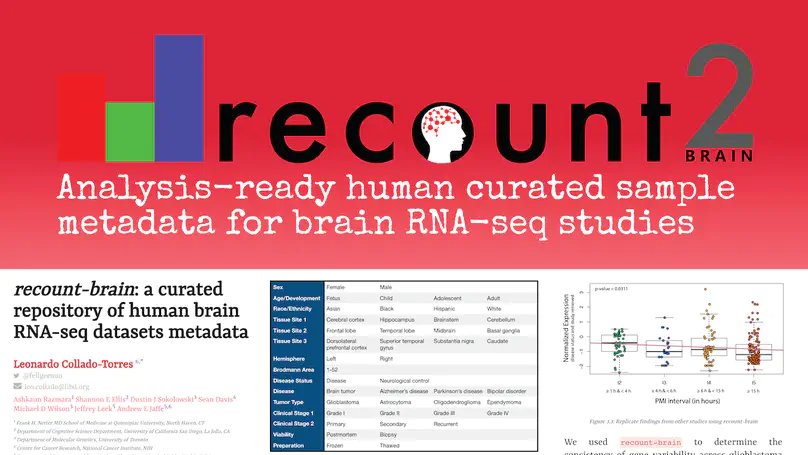
The usability of publicly-available gene expression data is often limited by the availability of high-quality, standardized biological phenotype and experimental condition information (“metadata”). We released the recount2 project, which involved re-processing ∼70,000 samples in the Sequencing Read Archive (SRA), Genotype-Tissue Expression (GTEx), and The Cancer Genome Atlas (TCGA) projects. While samples from the latter two projects are well-characterized with extensive metadata, the ∼50,000 RNA-seq samples from SRA in recount2 are inconsistently annotated with metadata. Tissue type, sex, and library type can be estimated from the RNA sequencing (RNA-seq) data itself. However, more detailed and harder to predict metadata, like age and diagnosis, must ideally be provided by labs that deposit the data. To facilitate more analyses within human brain tissue data, we have complemented phenotype predictions by manually constructing a uniformly-curated database of public RNA-seq samples present in SRA and recount2. We describe the reproducible curation process for constructing recount-brain that involves systematic review of the primary manuscript, which can serve as a guide to annotate other studies and tissues. We further expanded recount-brain by merging it with GTEx and TCGA brain samples as well as linking to controlled vocabulary terms for tissue, Brodmann area and disease. Furthermore, we illustrate how to integrate the sample metadata in recount-brain with the gene expression data in recount2 to perform differential expression analysis. We then provide three analysis examples involving modeling postmortem interval, glioblastoma, and meta-analyses across GTEx and TCGA. Overall, recount-brain facilitates expression analyses and improves their reproducibility as individual researchers do not have to manually curate the sample metadata. recount-brain is available via the add_metadata() function from the recount Bioconductor package at bioconductor.org/packages/recount.
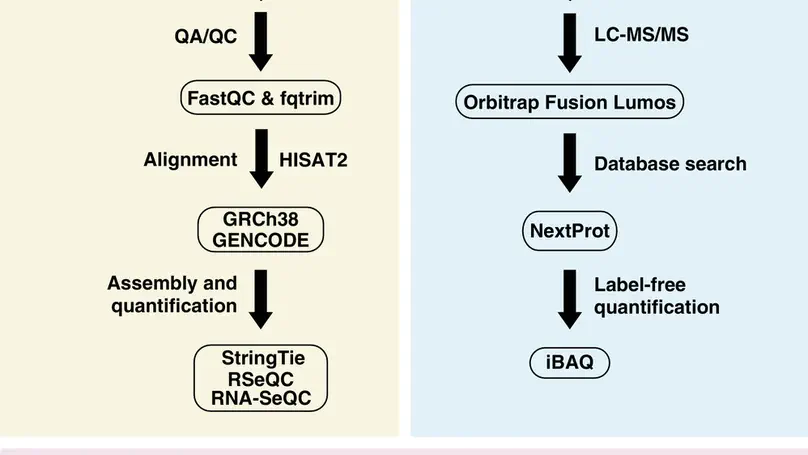
Understanding the molecular profile of every human cell type is essential for understanding its role in normal physi-ology and disease. Technological advancements in DNA sequencing, mass spectrometry, and computational methodsallow us to carry out multiomics analyses although such approaches are not routine yet. Human umbilical vein endothe-lial cells (HUVECs) are a widely used model system to study pathological and physiological processes associated withthe cardiovascular system. In this study, next-generation sequencing and high-resolution mass spectrometry to profilethe transcriptome and proteome of primary HUVECs is employed. Analysis of 145 million paired-end reads from next-generation sequencing confirmed expression of 12 186 protein-coding genes (FPKM >= 0.1), 439 novel long non-codingRNAs, and revealed 6089 novel isoforms that were not annotated in GENCODE. Proteomics analysis identifies 6477proteins including confirmation ofN-termini for 1091 proteins, isoforms for 149 proteins, and 1034 phosphosites. Adatabase search to specifically identify other post-translational modifications provide evidence for a number of modifi-cation sites on 117 proteins which include ubiquitylation, lysine acetylation, and mono-, di- and tri-methylation events.Evidence for 11 ‘missing proteins’, which are proteins for which there was insufficient or no protein level evidence, isprovided. Peptides supporting missing protein and novel events are validated by comparison of MS/MS fragmentationpatterns with synthetic peptides. Finally, 245 variant peptides derived from 207 expressed proteins in addition to alternatetranslational start sites for seven proteins and evidence for novel proteoforms for five proteins resulting from alternativesplicing are identified. Overall, it is believed that the integrated approach employed in this study is widely applicable tostudy any primary cell type for deeper molecular characterization.
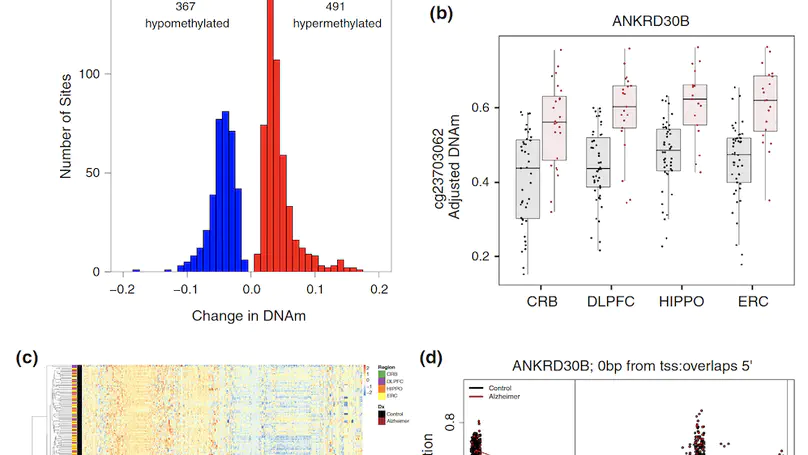
Late-onset Alzheimer’s disease (AD) is a complex age-related neurodegenerative disorder that likely involves epigenetic factors. To better understand the epigenetic state associated with AD, we surveyed 420,852 DNA methylation (DNAm) sites from neurotypical controls (N=49) and late-onset AD patients (N=24) across four brain regions (hippocampus, entorhinal cortex, dorsolateral prefrontal cortex and cerebellum). We identified 858 sites with robust differential methylation collectively annotated to 772 possible genes (FDR<5%, within 10 kb). These sites were overrepresented in AD genetic risk loci (p=0.00655) and were enriched for changes during normal aging (p<2.2×10−16), and nearby genes were enriched for processes related to cell-adhesion, immunity, and calcium homeostasis (FDR<5%). To functionally validate these associations, we generated and analyzed corresponding transcriptome data to prioritize 130 genes within 10 kb of the differentially methylated sites. These 130 genes were differentially expressed between AD cases and controls and their expression was associated with nearby DNAm (p<0.05). This integrated analysis implicates novel genes in Alzheimer’s disease, such as ANKRD30B. These results highlight DNAm differences in Alzheimer’s disease that have gene expression correlates, further implicating DNAm as an epigenetic mechanism underlying pathological molecular changes associated with AD. Furthermore, our framework illustrates the value of integrating epigenetic and transcriptomic data for understanding complex disease.
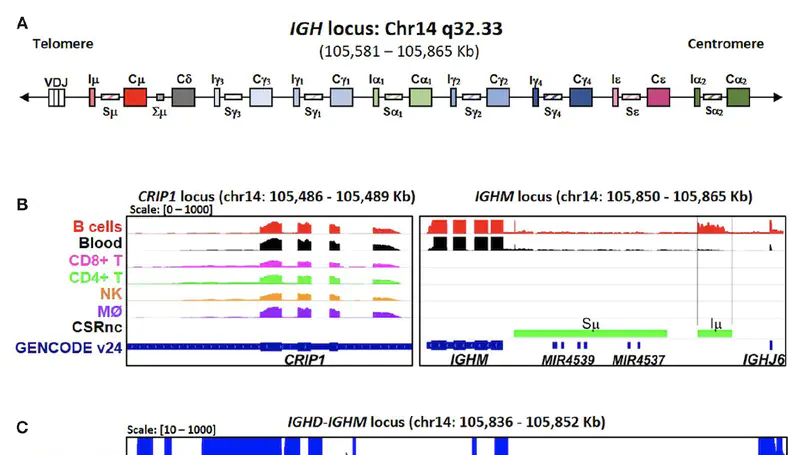
Background: Antibody class switch recombination (CSR) to IgG, IgA or IgE is a hallmark of adaptive immunity, allowing antibody function diversification beyond IgM. CSR involves a deletion of the IgM/IgD constant region genes placing a new acceptor Constant (CH) gene, downstream of the VDJH exon. CSR depends on non-coding (CSRnc) transcription of donor Iμ and acceptor IH exons, located 5′ upstream of each CH coding gene. Although our knowledge of the role of CSRnc transcription has advanced greatly, its extension and importance in healthy and diseased humans is scarce. Methods: We analyzed CSRnc transcription in 70,603 publicly available RNA-seq samples, including GTEx, TCGA and the Sequence Read Archive (SRA) using recount2, an online resource consisting of normalized RNA-seq gene and exon counts, as well as coverage BigWig files that can be programmatically accessed through R. CSRnc transcription was validated with a qRT-PCR assay for Iμ, Iγ1 and Iγ3 in humans in response to vaccination. Results: We mapped IH transcription for the human IgH locus, including the less understood IGHD gene. CSRnc transcription was restricted to B cells and is widely distributed in normal adult tissues, but predominant in blood, spleen, MALT-containing tissues, visceral adipose tissue and some so-called ‘immune privileged’ tissues. However, significant Iγ4 expression was found even in non-lymphoid fetal tissues. CSRnc expression in cancer tissues mimicked the expression of their normal counterparts, with notable pattern changes in some common cancer subsets. CSRnc transcription in tumors appears to result from tumor infiltration by B cells, since CSRnc transcription was not detected in corresponding tumor-derived immortal cell lines. Additionally, significantly increased Iδ transcription in ileal mucosa in Crohn’s disease with ulceration was found. Conclusions: CSRnc transcription occurs in multiple anatomical locations beyond classical secondary lymphoid organs, representing a potentially useful marker of effector B cell responses in normal and pathological immune responses. The pattern of IH exon expression may reveal clues of the local immune response (i.e. cytokine milieu) in health and disease. This is a great example of how the public recount2 data can be used to further our understanding of transcription, including regions outside the known transcriptome.
Genome-wide association studies have identified 108 schizophrenia risk loci, but biological mechanisms for individual loci are largely unknown. Using developmental, genetic and illness-based RNA sequencing expression analysis in human brain, we characterized the human brain transcriptome around these loci and found enrichment for developmentally regulated genes with novel examples of shifting isoform usage across pre- and postnatal life. We found widespread expression quantitative trait loci (eQTLs), including many with transcript specificity and previously unannotated sequence that were independently replicated. We leveraged this general eQTL database to show that 48.1% of risk variants for schizophrenia associate with nearby expression. We lastly found 237 genes significantly differentially expressed between patients and controls, which replicated in an independent dataset, implicated synaptic processes, and were strongly regulated in early development. These findings together offer genetics- and diagnosis-related targets for better modeling of schizophrenia risk. This resource is publicly available at http://eqtl.brainseq.org/phase1.
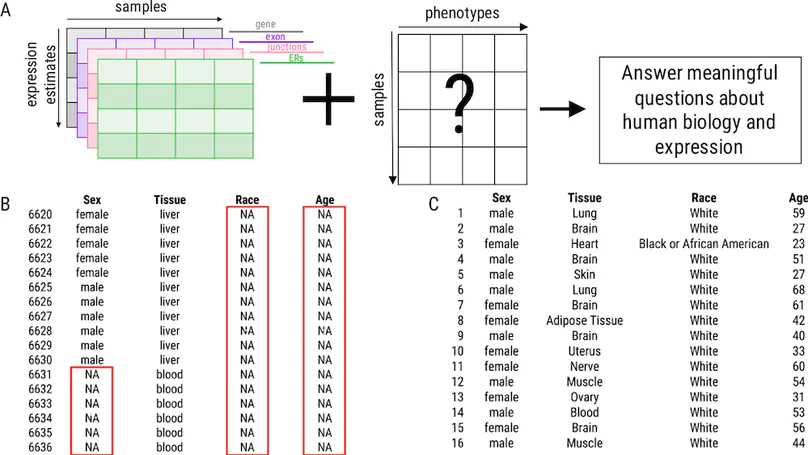
Background: Publicly available genomic data are a valuable resource for studying normal human variation and disease, but these data are often not well labeled or annotated. The lack of phenotype information for public genomic data severely limits their utility for addressing targeted biological questions. Results: We develop an in silico phenotyping approach for predicting critical missing annotation directly from genomic measurements using, well-annotated genomic and phenotypic data produced by consortia like TCGA and GTEx as training data. We apply in silico phenotyping to a set of 70,000 RNA-seq samples we recently processed on a common pipeline as part of the recount2 project (https://jhubiostatistics.shinyapps.io/recount/). We use gene expression data to build and evaluate predictors for both biological phenotypes (sex, tissue, sample source) and experimental conditions (sequencing strategy). We demonstrate how these predictions can be used to study cross-sample properties of public genomic data, select genomic projects with specific characteristics, and perform downstream analyses using predicted phenotypes. The methods to perform phenotype prediction are available in the phenopredict R package (https://github.com/leekgroup/phenopredict) and the predictions for recount2 are available from the recount R package (https://bioconductor.org/packages/release/bioc/html/recount.html). Conclusion: Having leveraging massive public data sets to generate a well-phenotyped set of expression data for more than 70,000 human samples, expression data is available for use on a scale that was not previously feasible.
More than 70,000 short-read RNA-sequencing samples are publicly available through the recount2 project, a curated database of summary coverage data. However, no current methods can estimate transcript-level abundances using the reduced-representation information stored in this database. Here we present a linear model utilizing coverage of junctions and subdivided exons to generate transcript abundance estimates of comparable accuracy to those obtained from methods requiring read-level data. Our approach flexibly models bias, produces standard errors, and is easy to refresh given updated annotation. We illustrate our method on simulated and real data and release transcript abundance estimates for the samples in recount2.
![recount workflow: Accessing over 70,000 human RNA-seq samples with Bioconductor [version 1; referees: 1 approved, 2 approved with reservations]](/publication/2017-08_recountworkflow/featured_hu3895338370ae321089968d378f88b52e_7454_808x455_fill_q75_h2_lanczos_smart1_3.webp)
The recount2 resource is composed of over 70,000 uniformly processed human RNA-seq samples spanning TCGA and SRA, including GTEx. The processed data can be accessed via the recount2 website and the recount Bioconductor package. This workflow explains in detail how to use the recount package and how to integrate it with other Bioconductor packages for several analyses that can be carried out with the recount2 resource. In particular, we describe how the coverage count matrices were computed in recount2 as well as different ways of obtaining public metadata, which can facilitate downstream analyses. Step-by-step directions show how to do a gene-level differential expression analysis, visualize base-level genome coverage data, and perform an analyses at multiple feature levels. This workflow thus provides further information to understand the data in recount2 and a compendium of R code to use the data.
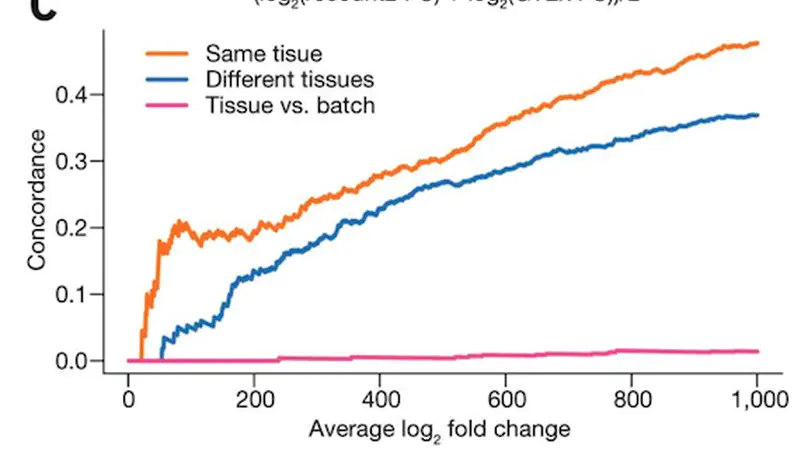
recount2 is a resource of processed and summarized expression data spanning over 70,000 human RNA-seq samples from the Sequence Read Archive (SRA). The associated recount Bioconductor package provides a convenient API for querying, downloading, and analyzing the data. Each processed study consists of meta/phenotype data, the expression levels of genes and their underlying exons and splice junctions, and corresponding genomic annotation. We also provide data summarization types for quantifying novel transcribed sequence including base-resolution coverage and potentially unannotated splice junctions. We present workflows illustrating how to use recount to perform differential expression analysis including meta-analysis, annotation-free base-level analysis, and replication of smaller studies using data from larger studies. recount provides a valuable and user-friendly resource of processed RNA-seq datasets to draw additional biological insights from existing public data. The resource is available at https://jhubiostatistics.shinyapps.io/recount/.
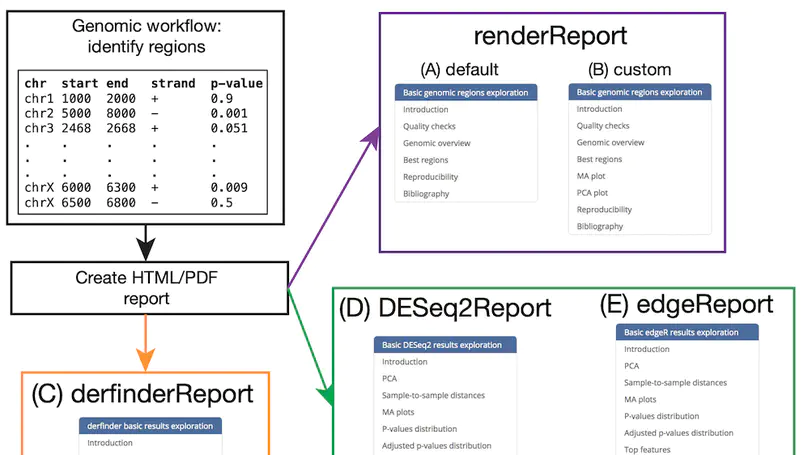
regionReport is an R package for generating detailed interactive reports from region-level genomic analyses as well as feature-level RNA-seq. The report includes quality-control checks, an overview of the results, an interactive table of the genomic regions or features of interest and reproducibility information. regionReport provides specialised reports for exploring DESeq2, edgeR, or derfinder differential expression analyses results. regionReport is also flexible and can easily be expanded with report templates for other analysis pipelines.
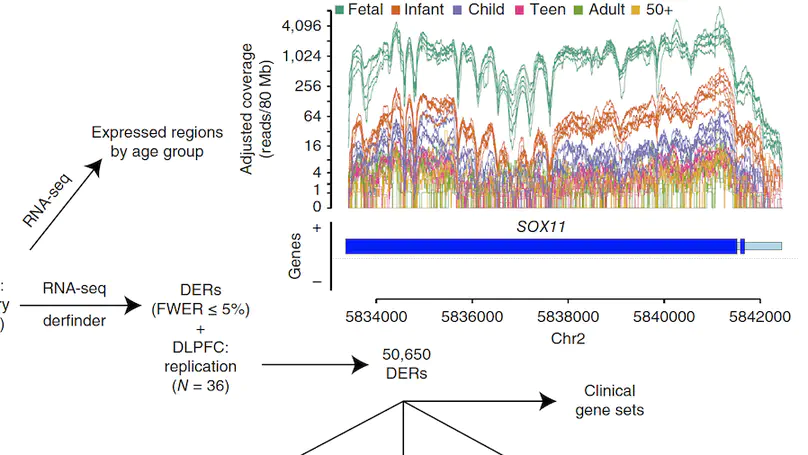
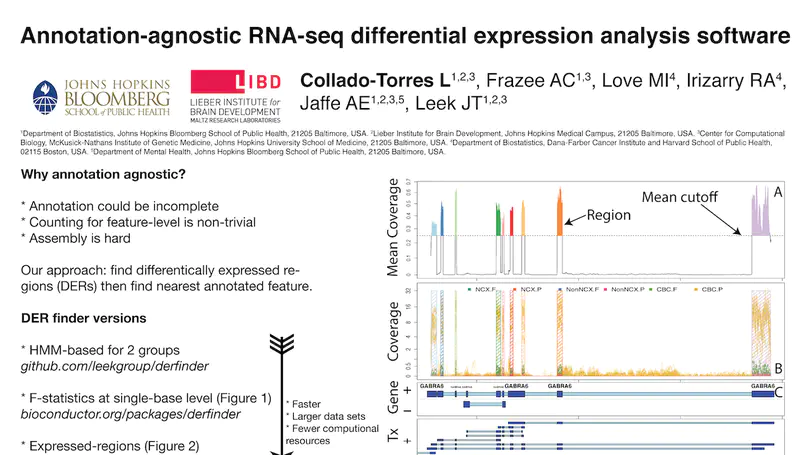
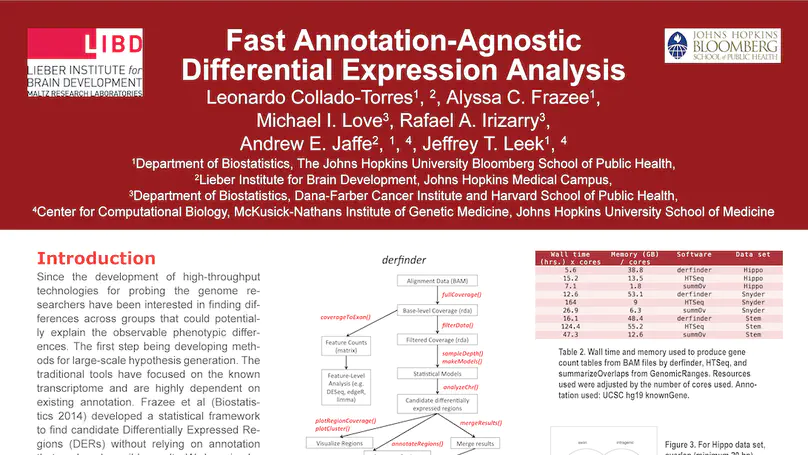
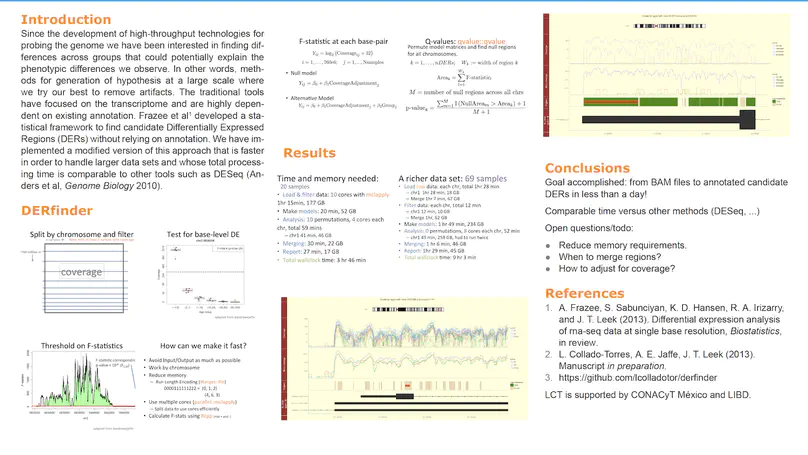
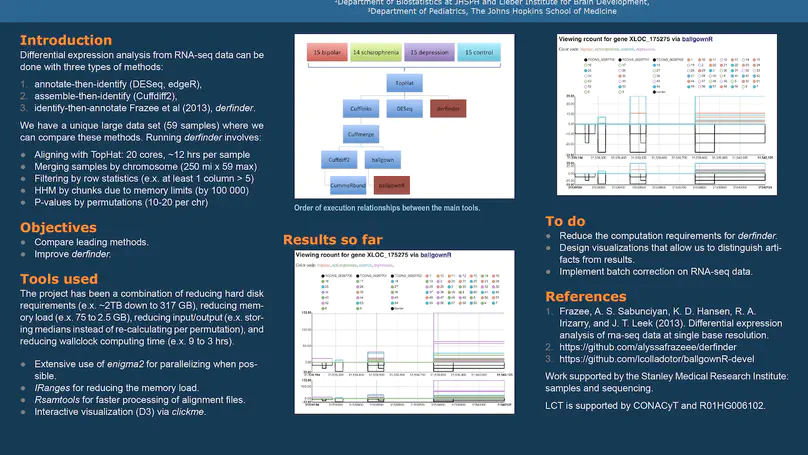
Transcriptome analysis of human brain provides fundamental insight into development and disease, but it largely relies on existing annotation. We sequenced transcriptomes of 72 prefrontal cortex samples across six life stages and identified 50,650 differentially expression regions (DERs) associated with developmental and aging, agnostic of annotation. While many DERs annotated to non-exonic sequence (41.1%), most were similarly regulated in cytosolic mRNA extracted from independent samples. The DERs were developmentally conserved across 16 brain regions and in the developing mouse cortex, and were expressed in diverse cell and tissue types. The DERs were further enriched for active chromatin marks and clinical risk for neurodevelopmental disorders such as schizophrenia. Lastly, we demonstrate quantitatively that these DERs associate with a changing neuronal phenotype related to differentiation and maturation. These data show conserved molecular signatures of transcriptional dynamics across brain development, have potential clinical relevance and highlight the incomplete annotation of the human brain transcriptome.
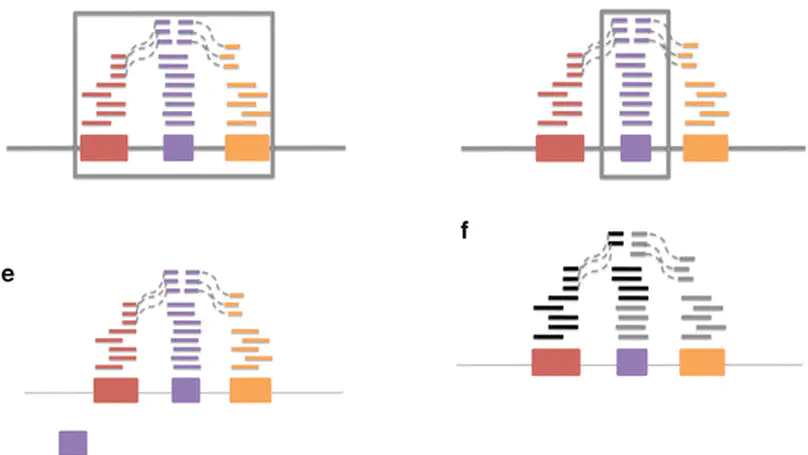
There has been a major shift from microarrays to RNA-sequencing (RNA-seq) for measuring gene expression as the price per measurement between these technologies has become comparable. The advantages of RNA-seq are increased measurement flexibility to detect alternative transcription, allele specific transcription, or transcription outside of known coding regions. The price of this increased flexibility is: (a) an increase in raw data size and (b) more decisions that must be made by the data analyst. Here we provide a selective review and extension of our previous work in attempting to measure variability in results due to different choices about how to summarize and analyze RNA-sequencing data. We discuss a standard model for gene expression measurements that breaks variability down into variation due to technology, biology, and measurement error. Finally, wee show the importance of gene model selection, normalization, and choice for statistical model on the ultimate results of an RNA-sequencing experiment.
Many different systems of bacterial interactions have been described. However, relatively few studies have explored how interactions between different microorganisms might influence bacterial development. To explore such interspecies interactions, we focused on Bacillus subtilis, which characteristically develops into matrix-producing cannibals before entering sporulation. We investigated whether organisms from the natural environment of B. subtilis—the soil—were able to alter the development of B.subtilis. To test this possibility, we developed a coculture microcolony screen in which we used fluorescent reporters to identify soil bacteria able to induce matrix production in B. subtilis. Most of the bacteria that influence matrix production in B. subtilis are members of the genus Bacillus, suggesting that such interactions may be predominantly with close relatives. The interactions we observed were mediated via two different mechanisms. One resulted in increased expression of matrix genes via the activation of a sensor histidine kinase, KinD. The second was kinase independent and conceivably functions by altering the relative subpopulations of B. subtilis cell types by preferentially killing noncannibals. These two mechanisms were grouped according to the inducing strain’s relatedness to B. subtilis. Our results suggest that bacteria preferentially alter their development in response to secreted molecules from closely related bacteria and do so using mechanisms that depend on the phylogenetic relatedness of the interacting bacteria.
RegulonDB (http://regulondb.ccg.unam.mx/) is the primary reference database of the best-known regulatory network of any free-living organism, that of Escherichia coli K-12. The major conceptual change since 3 years ago is an expanded biological context so that transcriptional regulation is now part of a unit that initiates with the signal and continues with the signal transduction to the core of regulation, modifying expression of the affected target genes responsible for the response. We call these genetic sensory response units, or Gensor Units. We have initiated their high-level curation, with graphic maps and superreactions with links to other databases. Additional connectivity uses expandable submaps. RegulonDB has summaries for every transcription factor (TF) and TF-binding sites with internal symmetry. Several DNA-binding motifs and their sizes have been redefined and relocated. In addition to data from the literature, we have incorporated our own information on transcription start sites (TSSs) and transcriptional units (TUs), obtained by using high-throughput whole-genome sequencing technologies. A new portable drawing tool for genomic features is also now available, as well as new ways to download the data, including web services, files for several relational database manager systems and text files including BioPAX format.